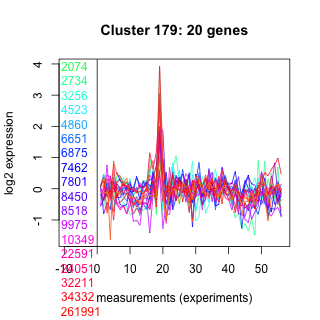
Thaps_hclust_0179 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006302 | double-strand break repair | 0.00144718 | 1 | Thaps_hclust_0179 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 0.00289256 | 1 | Thaps_hclust_0179 |
| GO:0009435 | NAD biosynthesis | 0.00433615 | 1 | Thaps_hclust_0179 |
| GO:0009190 | cyclic nucleotide biosynthesis | 0.031426 | 1 | Thaps_hclust_0179 |
| GO:0007242 | intracellular signaling cascade | 0.0592567 | 1 | Thaps_hclust_0179 |
| GO:0006457 | protein folding | 0.173725 | 1 | Thaps_hclust_0179 |
| GO:0008152 | metabolism | 0.459719 | 1 | Thaps_hclust_0179 |
|
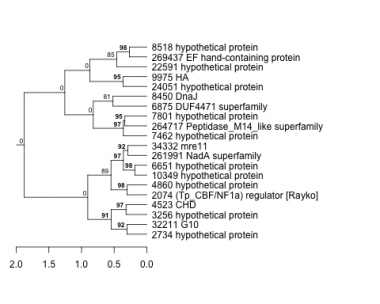
2074 : (Tp_CBF/NF1a) regulator [Rayko] |
6651 : hypothetical protein |
8518 : hypothetical protein |
32211 : G10 |
|
2734 : hypothetical protein |
6875 : DUF4471 superfamily |
9975 : HA |
34332 : mre11 |
|
3256 : hypothetical protein |
7462 : hypothetical protein |
10349 : hypothetical protein |
261991 : NadA superfamily |
|
4523 : CHD |
7801 : hypothetical protein |
22591 : hypothetical protein |
264717 : Peptidase_M14_like superfamily |
|
4860 : hypothetical protein |
8450 : DnaJ |
24051 : hypothetical protein |
269437 : EF hand-containing protein |
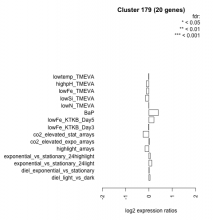
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| lowtemp_TMEVA | lowtemp_TMEVA | -0.012 | 0.975 |
| lowSi_TMEVA | lowSi_TMEVA | -0.146 | 0.974 |
| lowN_TMEVA | lowN_TMEVA | -0.004 | 1 |
| lowFe_TMEVA | lowFe_TMEVA | -0.106 | 0.786 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.219 | 0.284 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.013 | 0.968 |
| highpH_TMEVA | highpH_TMEVA | -0.112 | 0.52 |
| highlight_arrays | highlight_arrays | -0.165 | 0.32 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.124 | 0.759 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.055 | 0.645 |
| diel_light_vs_dark | diel_light_vs_dark | 0.050 | 0.875 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.013 | 0.964 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | -0.248 | 0.296 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.026 | 0.968 |
| BaP | BaP | 0.402 | 0.0602 |