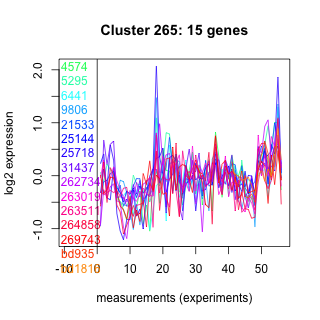
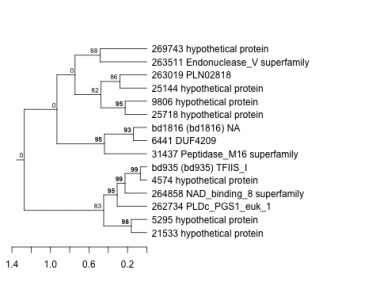
Thaps_hclust_0265 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006725 | aromatic compound metabolism | 0.0446037 | 1 | Thaps_hclust_0265 |
| GO:0006306 | DNA methylation | 0.0584155 | 1 | Thaps_hclust_0265 |
| GO:0006310 | DNA recombination | 0.0386288 | 1 | Thaps_hclust_0265 |
| GO:0006281 | DNA repair | 0.00379335 | 1 | Thaps_hclust_0265 |
| GO:0006260 | DNA replication | 0.0701108 | 1 | Thaps_hclust_0265 |
| GO:0006118 | electron transport | 0.432304 | 1 | Thaps_hclust_0265 |
| GO:0008152 | metabolism | 0.203212 | 1 | Thaps_hclust_0265 |
| GO:0006508 | proteolysis and peptidolysis | 0.459068 | 1 | Thaps_hclust_0265 |
|
4574 : hypothetical protein |
21533 : hypothetical protein |
262734 : PLDc_PGS1_euk_1 |
269743 : hypothetical protein |
|
5295 : hypothetical protein |
25144 : hypothetical protein |
263019 : PLN02818 |
bd935 : (bd935) TFIIS_I |
|
6441 : DUF4209 |
25718 : hypothetical protein |
263511 : Endonuclease_V superfamily |
bd1816 : (bd1816) NA |
|
9806 : hypothetical protein |
31437 : Peptidase_M16 superfamily |
264858 : NAD_binding_8 superfamily |
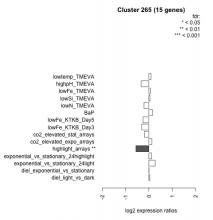
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | 0.042 | 0.915 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.302 | 0.276 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | -0.254 | 0.284 |
| BaP | BaP | 0.155 | 0.566 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.111 | 0.386 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | -0.192 | 0.468 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.090 | 0.787 |
| highpH_TMEVA | highpH_TMEVA | -0.334 | 0.0599 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.127 | 0.601 |
| lowFe_TMEVA | lowFe_TMEVA | 0.059 | 0.888 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.266 | 0.491 |
| lowN_TMEVA | lowN_TMEVA | -0.213 | 0.594 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.007 | 0.981 |
| lowSi_TMEVA | lowSi_TMEVA | 0.047 | 1 |
| highlight_arrays | highlight_arrays | -0.558 | 0.00157 |