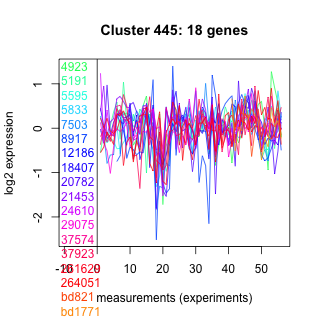
Thaps_hclust_0445 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | 0.00124018 | 1 | Thaps_hclust_0445 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.0313052 | 1 | Thaps_hclust_0445 |
| GO:0006457 | protein folding | 0.0784967 | 1 | Thaps_hclust_0445 |
|
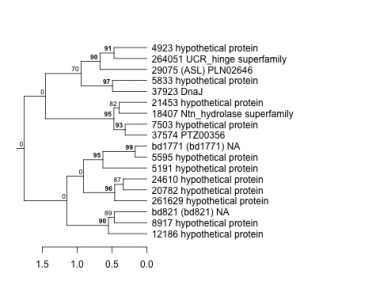
4923 : hypothetical protein |
8917 : hypothetical protein |
24610 : hypothetical protein |
261629 : hypothetical protein |
|
5191 : hypothetical protein |
12186 : hypothetical protein |
29075 : (ASL) PLN02646 |
264051 : UCR_hinge superfamily |
|
5595 : hypothetical protein |
18407 : Ntn_hydrolase superfamily |
37574 : PTZ00356 |
bd821 : (bd821) NA |
|
5833 : hypothetical protein |
20782 : hypothetical protein |
37923 : DnaJ |
bd1771 : (bd1771) NA |
|
7503 : hypothetical protein |
21453 : hypothetical protein |
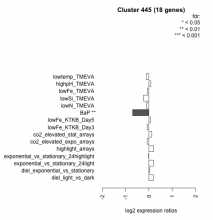
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| BaP | BaP | -0.696 | 0.00132 |
| lowSi_TMEVA | lowSi_TMEVA | -0.234 | 0.832 |
| lowtemp_TMEVA | lowtemp_TMEVA | -0.116 | 0.695 |
| lowN_TMEVA | lowN_TMEVA | -0.105 | 0.789 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | -0.074 | 0.833 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.071 | 0.81 |
| lowFe_TMEVA | lowFe_TMEVA | -0.058 | 0.888 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.010 | 0.957 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.031 | 0.896 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.071 | 0.767 |
| highpH_TMEVA | highpH_TMEVA | 0.080 | 0.686 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.121 | 0.66 |
| diel_light_vs_dark | diel_light_vs_dark | 0.190 | 0.433 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.199 | 0.604 |
| highlight_arrays | highlight_arrays | 0.205 | 0.232 |