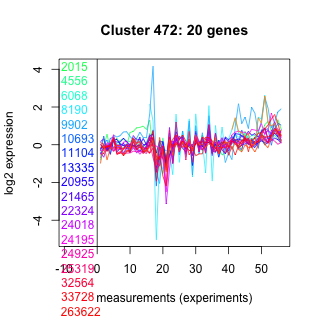
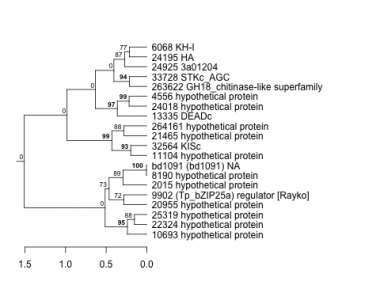
Thaps_hclust_0472 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0005975 | carbohydrate metabolism | 0.117597 | 1 | Thaps_hclust_0472 |
| GO:0006810 | transport | 0.266912 | 1 | Thaps_hclust_0472 |
| GO:0006468 | protein amino acid phosphorylation | 0.302703 | 1 | Thaps_hclust_0472 |
| GO:0006118 | electron transport | 0.364179 | 1 | Thaps_hclust_0472 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.40843 | 1 | Thaps_hclust_0472 |
| GO:0008152 | metabolism | 0.505245 | 1 | Thaps_hclust_0472 |
| GO:0007018 | microtubule-based movement | 0.068993 | 1 | Thaps_hclust_0472 |
| GO:0000074 | regulation of cell cycle | 0.0950978 | 1 | Thaps_hclust_0472 |
|
2015 : hypothetical protein |
10693 : hypothetical protein |
22324 : hypothetical protein |
32564 : KISc |
|
4556 : hypothetical protein |
11104 : hypothetical protein |
24018 : hypothetical protein |
33728 : STKc_AGC |
|
6068 : KH-I |
13335 : DEADc |
24195 : HA |
263622 : GH18_chitinase-like superfamily |
|
8190 : hypothetical protein |
20955 : hypothetical protein |
24925 : 3a01204 |
264161 : hypothetical protein |
|
9902 : (Tp_bZIP25a) regulator [Rayko] |
21465 : hypothetical protein |
25319 : hypothetical protein |
bd1091 : (bd1091) NA |
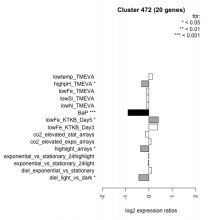
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | -0.432 | 0.0306 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.380 | 0.0967 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.405 | 0.0344 |
| BaP | BaP | -0.898 | 0.00037 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.008 | 0.96 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | -0.144 | 0.574 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.153 | 0.565 |
| highpH_TMEVA | highpH_TMEVA | -0.313 | 0.0494 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.094 | 0.739 |
| lowFe_TMEVA | lowFe_TMEVA | -0.094 | 0.813 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | -0.025 | 0.953 |
| lowN_TMEVA | lowN_TMEVA | -0.106 | 0.784 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.113 | 0.583 |
| lowSi_TMEVA | lowSi_TMEVA | -0.098 | 0.996 |
| highlight_arrays | highlight_arrays | -0.359 | 0.0203 |