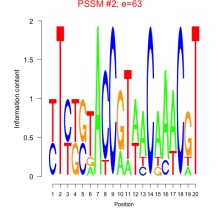
Phatr_bicluster_0008 Residual: 0.47
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0008 | 0.47 | Phaeodactylum tricornutum |
Displaying 1 - 28 of 28
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12510 protein phosphatase 2c (PP2Cc)
PHATRDRAFT_16648 ferric reductase-like protein (Got1 superfamily)
PHATRDRAFT_18228 triosephosphate isomerase (TIM)
PHATRDRAFT_34681 (NADB_Rossmann superfamily)
PHATRDRAFT_38448 m17 (leucyl aminopeptidase) family (Peptidase_M17 superfamily)
PHATRDRAFT_38498 sbf-domain containing protein (DUF4137)
PHATRDRAFT_44327 small-conductance mechanosensitive channel (MS_channel superfamily)
PHATRDRAFT_45608 dtw domain protein (DTW superfamily)
PHATRDRAFT_46353 phosphoglycerate bisphosphoglycerate mutase family protein (HP_PGM_like)
PHATRDRAFT_47106 m17 (leucyl aminopeptidase) family (Peptidase_M17 superfamily)
PHATRDRAFT_47293 thiamine monophosphate synthase (HMPP_kinase)
PHATRDRAFT_48726 elicitin-like protein sol8
PHATRDRAFT_7957 general transcription factor (Bromodomain)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009228 | thiamin biosynthesis | 0.00444005 | 1 | Phatr_bicluster_0008 |
| GO:0016192 | vesicle-mediated transport | 0.0191218 | 1 | Phatr_bicluster_0008 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0278439 | 1 | Phatr_bicluster_0008 |
| GO:0006508 | proteolysis and peptidolysis | 0.0394266 | 1 | Phatr_bicluster_0008 |
| GO:0008152 | metabolism | 0.364015 | 1 | Phatr_bicluster_0008 |


Comments