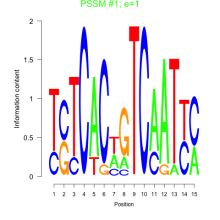
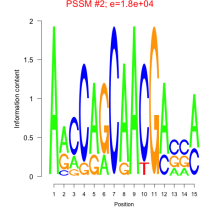
Phatr_bicluster_0016 Residual: 0.47
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0016 | 0.47 | Phaeodactylum tricornutum |
Displaying 1 - 34 of 34
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10479 plant adhesion molecule (RabGAP-TBC)
PHATRDRAFT_15495 glucokinase (Glucokinase superfamily)
PHATRDRAFT_16955 sodium symporter-related (NnrU superfamily)
PHATRDRAFT_35830 d-isomer specific 2-hydroxyacidnad-binding (FDH_GDH_like superfamily)
PHATRDRAFT_36098 selenium binding
PHATRDRAFT_40880 tpr domain protein
PHATRDRAFT_42577 agmatine ureohydrolase (Agmatinase-like)
PHATRDRAFT_42790 (COG5022)
PHATRDRAFT_44074 (ANK)
PHATRDRAFT_45345 zinc finger protein (RING)
PHATRDRAFT_47807 pentatricopeptiderepeat-containing protein
PHATRDRAFT_48851 (PAP_fibrillin superfamily)
PHATRDRAFT_49964 synoviolin 1 (HRD1)
PHATRDRAFT_50026 dd104upregulated upon bacterial challenge and trauma
PHATRDRAFT_50045 na+ proline symporter (SLC5-6-like_sbd superfamily)
PHATRDRAFT_50327 (GAL4)
PHATRDRAFT_50622 novel protein (zgc:64188)
PHATRDRAFT_52619 heat shock transcription factor 4 isoform 1 (HSF_DNA-bind)
PHATRDRAFT_54048 uric acid-xanthine permease (ncs2)
PHATRDRAFT_6940 tubby like protein 1 (Tub superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006527 | arginine catabolism | 0.00591934 | 1 | Phatr_bicluster_0016 |
| GO:0016567 | protein ubiquitination | 0.018343 | 1 | Phatr_bicluster_0016 |
| GO:0006564 | L-serine biosynthesis | 0.0234852 | 1 | Phatr_bicluster_0016 |
| GO:0006096 | glycolysis | 0.0990654 | 1 | Phatr_bicluster_0016 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.134605 | 1 | Phatr_bicluster_0016 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.143861 | 1 | Phatr_bicluster_0016 |
| GO:0006118 | electron transport | 0.144823 | 1 | Phatr_bicluster_0016 |
| GO:0006412 | protein biosynthesis | 0.385887 | 1 | Phatr_bicluster_0016 |
| GO:0006810 | transport | 0.440472 | 1 | Phatr_bicluster_0016 |

Comments