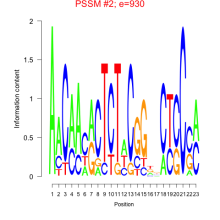
Phatr_bicluster_0019 Residual: 0.43
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0019 | 0.43 | Phaeodactylum tricornutum |
Displaying 1 - 29 of 29
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10508 af436836_1 at5g35180 t25c13_60 (DUF1336 superfamily)
PHATRDRAFT_10704 cyclin dependent kinase c (STKc_CDK9_like)
PHATRDRAFT_11414 mitochondrial phosphate carrier protein (Mito_carr)
PHATRDRAFT_13192 cdc5 cell division cycle 5-like (SANT)
PHATRDRAFT_14090 nuclear accommodation of mitochondria (AAA_11)
PHATRDRAFT_14771 interferon-induced mx protein (DLP_1)
PHATRDRAFT_14983 dead box atp-dependent rna helicase (DEADc)
PHATRDRAFT_15217 d-isomer specific 2-hydroxyacidnad-binding (2-Hacid_dh_C superfamily)
PHATRDRAFT_21988 (PRK09279)
PHATRDRAFT_22404 pyruvate kinase (PRK06247)
PHATRDRAFT_31364 trna-dihydrouridine synthase (DUS_like_FMN)
PHATRDRAFT_33458 (Trp-synth-beta_II superfamily)
PHATRDRAFT_38120 (Glyco_tranf_GTA_type superfamily)
PHATRDRAFT_38270 3-5 exonuclease (DnaQ_like_exo superfamily)
PHATRDRAFT_39832 glycogenin-like protein (Glyco_tranf_GTA_type superfamily)
PHATRDRAFT_45758 (CysJ)
PHATRDRAFT_46160 protein phosphatase (PP2Cc)
PHATRDRAFT_47020 cytokine induced apoptosis inhibitorisoform cra_b (CIAPIN1)
PHATRDRAFT_48732 prolyl 4-hydroxylase alpha subunit (2OG-FeII_Oxy_3 superfamily)
PHATRDRAFT_49205 jun d proto-oncogene (B_zip1)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0016051 | carbohydrate biosynthesis | 0.00886476 | 1 | Phatr_bicluster_0019 |
| GO:0016310 | phosphorylation | 0.0307218 | 1 | Phatr_bicluster_0019 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0414888 | 1 | Phatr_bicluster_0019 |
| GO:0006096 | glycolysis | 0.0752324 | 1 | Phatr_bicluster_0019 |
| GO:0019538 | protein metabolism | 0.0773059 | 1 | Phatr_bicluster_0019 |
| GO:0006468 | protein amino acid phosphorylation | 0.323667 | 1 | Phatr_bicluster_0019 |
| GO:0006810 | transport | 0.352949 | 1 | Phatr_bicluster_0019 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.394049 | 1 | Phatr_bicluster_0019 |
| GO:0006118 | electron transport | 0.408154 | 1 | Phatr_bicluster_0019 |


Comments