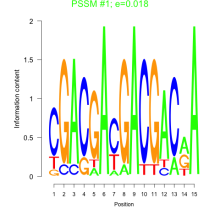
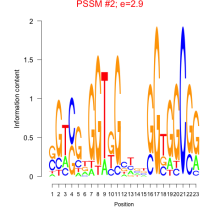
Phatr_bicluster_0024 Residual: 0.42
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0024 | 0.42 | Phaeodactylum tricornutum |
Displaying 1 - 37 of 37
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10567 short chain dehydrogenase (NADB_Rossmann superfamily)
PHATRDRAFT_12586 ribosomal large subunit pseudouridine synthase d (RluA)
PHATRDRAFT_14374 mbf1_canal multiprotein-bridging factor 1 (HTH_XRE)
PHATRDRAFT_15294 trna (5-methylaminomethyl-2-thiouridylate)-methyltransferase (tRNA_Me_trans)
PHATRDRAFT_15777 ferredoxin-nadp oxidoreductase (PLN03116)
PHATRDRAFT_18793 tnf receptor-associated protein (PRK05218)
PHATRDRAFT_21744 calmodulin (PTZ00184)
PHATRDRAFT_22873 atp adp translocator (PTZ00169)
PHATRDRAFT_30770 af503284_1microsomal cytochrome b5 (Cyt-b5)
PHATRDRAFT_33159 26s proteasome regulatory chain 4 (PTZ00361)
PHATRDRAFT_37556 transcription factorcyclin-related
PHATRDRAFT_38526 gliotoxin biosynthesis protein
PHATRDRAFT_39528 (CPSaseII_lrg)
PHATRDRAFT_39710 sam-dependent methyltransferase (Methyltransf_31)
PHATRDRAFT_41031 sec61 signal peptide plus 9 transmembrane domain-containing protein (PTZ00219)
PHATRDRAFT_41417 heat shock protein (NBD_sugar-kinase_HSP70_actin superfamily)
PHATRDRAFT_42830 3hc4 type family protein (RING)
PHATRDRAFT_42867 (LAM superfamily)
PHATRDRAFT_43251 adp-ribosylation factor (Arf1_5_like)
PHATRDRAFT_44132 aspartyl asparaginyl beta-hydroxylase (Asp_Arg_Hydrox superfamily)
PHATRDRAFT_47402 sym1_neucr protein sym-1 (Mpv17_PMP22)
PHATRDRAFT_47831 egf-like repeats and discoidin i-like domains 3
PHATRDRAFT_48094 proteophosphoglycan ppg4
PHATRDRAFT_49892 (DUF1997 superfamily)
PHATRDRAFT_50006 phytochelatin synthase (Phytochelatin superfamily)
PHATRDRAFT_50914 xenobiotic reductase b (OYE_like_FMN)
PHATRDRAFT_51604 swi2 snf2-like protein (SNF2_N)
PHATRDRAFT_54960 (SPFH_prohibitin)
PHATRDRAFT_55086 delta-1-pyrroline 5-carboxylase synthetase (PLN02418)
PHATRDRAFT_5512 alanine racemase family protein (PLPDE_III_YBL036c_euk)
PHATRDRAFT_5902 heme oxygenase (HemeO)
PHATRDRAFT_769 nuclear vcp-like protein (SpoVK)
PHATRDRAFT_8269 rnp-1 like rna-binding protein (RRM_SF superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009306 | protein secretion | 0.01133 | 1 | Phatr_bicluster_0024 |
| GO:0006788 | heme oxidation | 0.0169488 | 1 | Phatr_bicluster_0024 |
| GO:0019856 | pyrimidine base biosynthesis | 0.0169488 | 1 | Phatr_bicluster_0024 |
| GO:0018193 | peptidyl-amino acid modification | 0.028095 | 1 | Phatr_bicluster_0024 |
| GO:0006526 | arginine biosynthesis | 0.0391202 | 1 | Phatr_bicluster_0024 |
| GO:0009401 | phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.055434 | 1 | Phatr_bicluster_0024 |
| GO:0006352 | transcription initiation | 0.0661623 | 1 | Phatr_bicluster_0024 |
| GO:0006807 | nitrogen compound metabolism | 0.076774 | 1 | Phatr_bicluster_0024 |
| GO:0008033 | tRNA processing | 0.107921 | 1 | Phatr_bicluster_0024 |
| GO:0007264 | small GTPase mediated signal transduction | 0.113014 | 1 | Phatr_bicluster_0024 |

Comments