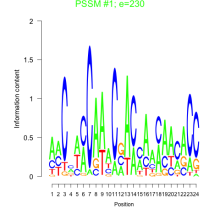
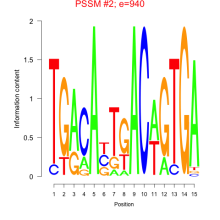
Phatr_bicluster_0035 Residual: 0.39
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0035 | 0.39 | Phaeodactylum tricornutum |
Displaying 1 - 43 of 43
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_1372 exg_blugrglucan-beta-glucosidase precursor (exo--beta-glucanase) (BglC)
PHATRDRAFT_17555 mitochondrial carrier protein (Mito_carr)
PHATRDRAFT_18096 dnaj domain (DnaJ)
PHATRDRAFT_21660 (PIKKc_TOR)
PHATRDRAFT_34741 thioredoxin family protein (Thioredoxin_like superfamily)
PHATRDRAFT_40891 muc17 protein (PHA03247)
PHATRDRAFT_42794 tesmin tso1-like cxc domain containing protein (TCR)
PHATRDRAFT_43132 (RING)
PHATRDRAFT_43291 adaptor protein kanadaptin (FHA)
PHATRDRAFT_43493 (Dynactin_p62 superfamily)
PHATRDRAFT_43944 nadh dehydrogenase (PLN03132)
PHATRDRAFT_44181 (FHA)
PHATRDRAFT_44334 pseudouridine synthase family protein (PseudoU_synth_RluCD_like)
PHATRDRAFT_44390 (SnoaL_2)
PHATRDRAFT_44700 jun protein (BRLZ)
PHATRDRAFT_45033 early transcribed membrane protein
PHATRDRAFT_45096 (EEP superfamily)
PHATRDRAFT_45293 (SET)
PHATRDRAFT_45435 ppr protein (PPR_2)
PHATRDRAFT_46470 (BAR)
PHATRDRAFT_46577 pseudouridylate synthase (PseudoU_synth_RluCD_like)
PHATRDRAFT_46896 prolyl 4-hydroxylase alpha subunit-like2og-feoxygenase family protein (2OG-FeII_Oxy_3)
PHATRDRAFT_48054 epsilon frustilin
PHATRDRAFT_48570 rhodanese domain protein (RHOD)
PHATRDRAFT_49160 (ARM)
PHATRDRAFT_50364 sjchgc08052 protein
PHATRDRAFT_50425 (Nsp1_C superfamily)
PHATRDRAFT_50615 (Sec7)
PHATRDRAFT_50825 ruvb2_ustma-like helicase 2 (TIP49)
PHATRDRAFT_54558 epsilon frustilin
PHATRDRAFT_8860 lecithin:cholesterol acyltransferase acyl-ceramide synthase (PLN02517 superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 0.0103397 | 1 | Phatr_bicluster_0035 |
| GO:0006396 | RNA processing | 0.0153703 | 1 | Phatr_bicluster_0035 |
| GO:0006629 | lipid metabolism | 0.0605653 | 1 | Phatr_bicluster_0035 |
| GO:0007018 | microtubule-based movement | 0.0990303 | 1 | Phatr_bicluster_0035 |
| GO:0019538 | protein metabolism | 0.117709 | 1 | Phatr_bicluster_0035 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.120788 | 1 | Phatr_bicluster_0035 |
| GO:0006810 | transport | 0.139001 | 1 | Phatr_bicluster_0035 |
| GO:0006118 | electron transport | 0.185759 | 1 | Phatr_bicluster_0035 |
| GO:0016567 | protein ubiquitination | 0.222443 | 1 | Phatr_bicluster_0035 |
| GO:0006457 | protein folding | 0.337129 | 1 | Phatr_bicluster_0035 |

Comments