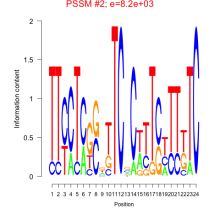
Phatr_bicluster_0041 Residual: 0.40
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0041 | 0.40 | Phaeodactylum tricornutum |
Displaying 1 - 35 of 35
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10208 phosphoribulokinase precursor (PRK)
PHATRDRAFT_10261 beta-ig-h3 fasciclin (Fasciclin)
PHATRDRAFT_14682 peptidyl-prolyl cis-transcyclophilin type (cyclophilin)
PHATRDRAFT_16481 light harvesting complex protein (Chloroa_b-bind)
PHATRDRAFT_32261 possible gram-negative pili assembly chaperone (PAP_fibrillin superfamily)
PHATRDRAFT_34177 rubredoxin familyexpressed
PHATRDRAFT_34584 (TRX_Fd_family)
PHATRDRAFT_38893 mgc86224 protein (ESCRT-II superfamily)
PHATRDRAFT_39940 tellurite resistance protein (AdoMet_MTases)
PHATRDRAFT_41079 (Cupin_7)
PHATRDRAFT_41243 gamma-butyrobetaine hydroxylase (CAS_like superfamily)
PHATRDRAFT_43208 vegetatible incompatibility protein (WD40 superfamily)
PHATRDRAFT_43685 ax110p-like protein (GFO_IDH_MocA superfamily)
PHATRDRAFT_43828 ion transport protein (Ion_trans)
PHATRDRAFT_44955 4-hydroxy-3-methylbut-2-en-1-yl diphosphatechloroplast precursor (1-hydroxy-2-methyl-2--butenyl 4-diphosphate synthase) (PLN02925)
PHATRDRAFT_46657 ucria_soltucytochrome b6-f complex iron-sulfurchloroplast precursor (rieske iron-sulfur protein) (plastohydroquinone:plastocyanin oxidoreductase iron-sulfur protein) (PRK13474)
PHATRDRAFT_47426 r 9 protein (DUF760)
PHATRDRAFT_49151 conserved protein (NADB_Rossmann superfamily)
PHATRDRAFT_49777 aldo keto reductase (Aldo_ket_red)
PHATRDRAFT_49795 (Abhydrolase_5)
PHATRDRAFT_54015 at4g37930 f20d10_50 (SHMT)
PHATRDRAFT_54731 thylakoid ascorbate peroxidase (ascorbate_peroxidase)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009058 | biosynthesis | 0.115816 | 1 | Phatr_bicluster_0041 |
| GO:0005975 | carbohydrate metabolism | 0.158622 | 1 | Phatr_bicluster_0041 |
| GO:0006812 | cation transport | 0.0710389 | 1 | Phatr_bicluster_0041 |
| GO:0007155 | cell adhesion | 0.0347981 | 1 | Phatr_bicluster_0041 |
| GO:0006118 | electron transport | 0.125163 | 1 | Phatr_bicluster_0041 |
| GO:0006811 | ion transport | 0.0295173 | 1 | Phatr_bicluster_0041 |
| GO:0009765 | photosynthesis light harvesting | 0.101113 | 1 | Phatr_bicluster_0041 |
| GO:0006457 | protein folding | 0.275986 | 1 | Phatr_bicluster_0041 |
| GO:0016114 | terpenoid biosynthesis | 0.00542673 | 1 | Phatr_bicluster_0041 |
| GO:0006810 | transport | 0.412691 | 1 | Phatr_bicluster_0041 |

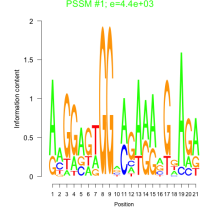
Comments