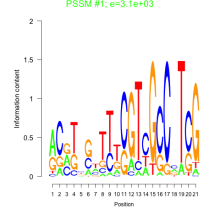
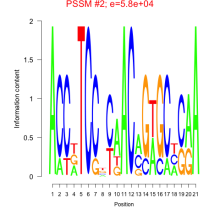
Phatr_bicluster_0056 Residual: 0.30
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0056 | 0.30 | Phaeodactylum tricornutum |
Displaying 1 - 31 of 31
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_14177 neural precursor celldevelopmentally down-regulated 4 (HECTc)
PHATRDRAFT_15207 wd repeat domain 45 (COG2319)
PHATRDRAFT_1534 at5g47240 mql5_10 (Nudix_Hydrolase superfamily)
PHATRDRAFT_15341 anion exchange family protein (HCO3_cotransp superfamily)
PHATRDRAFT_15929 at5g19750 t29j13_170 (Mpv17_PMP22)
PHATRDRAFT_16014 protein kinase domain containingexpressed (S_TKc)
PHATRDRAFT_16105 rna binding trna methyltransferase (tRNA_m1G_MT)
PHATRDRAFT_16471 transmembrane proteinisoform cra_a (Solute_trans_a)
PHATRDRAFT_21996 iron-sulfur cluster assembly protein (COG0694)
PHATRDRAFT_3834 nonsense-mediated mrna decay protein (NMD3)
PHATRDRAFT_39303 protein phosphatase 2c containing protein (PP2Cc)
PHATRDRAFT_44112 serinelong chain base subunit 1 (PLN03227)
PHATRDRAFT_46277 enhanced disease susceptibility salicylic acid induction deficient (MATE_like superfamily)
PHATRDRAFT_46830 dhhc zinc finger domain containing protein (zf-DHHC)
PHATRDRAFT_47669 delta 5 fatty acid desaturase d5 (Delta6-FADS-like)
PHATRDRAFT_48117 (zf-MYND)
PHATRDRAFT_48267 ntp7b_xenlasodium bile acid cotransporter 7-b (na(+) bile acid cotransporter 7-b) (solute carrier family 10 member 7-b) (DUF4137)
PHATRDRAFT_49592 (MFS)
PHATRDRAFT_49601 (LmjF365940-deam)
PHATRDRAFT_49646 serine-pyruvate aminotransferase (COG0075)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006636 | fatty acid desaturation | 0.0220309 | 1 | Phatr_bicluster_0056 |
| GO:0006820 | anion transport | 0.026385 | 1 | Phatr_bicluster_0056 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0414888 | 1 | Phatr_bicluster_0056 |
| GO:0006512 | ubiquitin cycle | 0.0814404 | 1 | Phatr_bicluster_0056 |
| GO:0009058 | biosynthesis | 0.095782 | 1 | Phatr_bicluster_0056 |
| GO:0006464 | protein modification | 0.13175 | 1 | Phatr_bicluster_0056 |
| GO:0008152 | metabolism | 0.134877 | 1 | Phatr_bicluster_0056 |
| GO:0006468 | protein amino acid phosphorylation | 0.323667 | 1 | Phatr_bicluster_0056 |

Comments