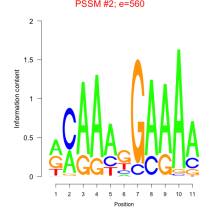
Phatr_bicluster_0058 Residual: 0.36
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0058 | 0.36 | Phaeodactylum tricornutum |
Displaying 1 - 46 of 46
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12788 mitochondrial carrier protein (Mito_carr)
PHATRDRAFT_14002 glycosyl transferase family 1 protein (PLN02275)
PHATRDRAFT_14937 carbon dioxide concentrating mechanism protein (TIGR00275)
PHATRDRAFT_15801 (DUF1349 superfamily)
PHATRDRAFT_15935 (NAD_binding_8 superfamily)
PHATRDRAFT_23830 phosphate-repressible phosphate permease-like protein (PHO4)
PHATRDRAFT_31339 (p450 superfamily)
PHATRDRAFT_37525 mbp-1 interacting protein-2a (Sedlin_N superfamily)
PHATRDRAFT_38899 sulfate transporter
PHATRDRAFT_40063 seed maturation proteinexpressed (DUF4149)
PHATRDRAFT_41265 tructure of aminoadipate-semialdehyde dehydrogenase- phosphopantetheinyl transferase (ACPS)
PHATRDRAFT_44003 aspartoacylase (PRK02259 superfamily)
PHATRDRAFT_44492 amino-acid n-acetyltransferase (AAK superfamily)
PHATRDRAFT_46926 aaa+-type atpase (DUF1995 superfamily)
PHATRDRAFT_47706 protein phosphatase 4 (formerly x)catalytic subunit (Arv1 superfamily)
PHATRDRAFT_47970 peptidyl-prolyl cis-transfkbp-type (FKBP_C)
PHATRDRAFT_48135 (PHD)
PHATRDRAFT_48282 (E_set_GO_C)
PHATRDRAFT_49223 periplasmic serine protease do (htra-1) (degP_htrA_DO)
PHATRDRAFT_49415 cobalamin synthesisp47k (COG0523)
PHATRDRAFT_49836 gpi transamidase component gpi16 subunit family protein (Gpi16 superfamily)
PHATRDRAFT_50149 cg3747-isoform a (SDF)
PHATRDRAFT_7195 zinc-binding hydrolase (nucleoside_deaminase)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006835 | dicarboxylic acid transport | 0.00641182 | 1 | Phatr_bicluster_0058 |
| GO:0006817 | phosphate transport | 0.0222757 | 1 | Phatr_bicluster_0058 |
| GO:0006633 | fatty acid biosynthesis | 0.0410037 | 1 | Phatr_bicluster_0058 |
| GO:0006810 | transport | 0.1228 | 1 | Phatr_bicluster_0058 |
| GO:0006508 | proteolysis and peptidolysis | 0.159337 | 1 | Phatr_bicluster_0058 |
| GO:0006118 | electron transport | 0.165064 | 1 | Phatr_bicluster_0058 |
| GO:0008152 | metabolism | 0.242573 | 1 | Phatr_bicluster_0058 |
| GO:0006457 | protein folding | 0.317339 | 1 | Phatr_bicluster_0058 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.515177 | 1 | Phatr_bicluster_0058 |

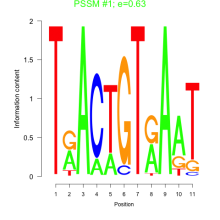
Comments