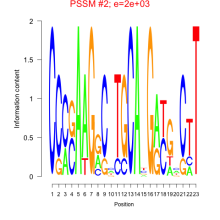
Phatr_bicluster_0063 Residual: 0.36
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0063 | 0.36 | Phaeodactylum tricornutum |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12990 dead-box protein abstrakt (PRK10590)
PHATRDRAFT_16120 cytoplasmic protein of eukaryotic origin-like (DUF2419 superfamily)
PHATRDRAFT_23306 vacuolar protein sorting 13b (VacB)
PHATRDRAFT_25 (COG1204)
PHATRDRAFT_267 insulin degrading enzyme (Ptr)
PHATRDRAFT_33266 solute carrier family 34 (sodium phosphate)member 1 (2a58)
PHATRDRAFT_36016 homology subfamily a member 5 (DnaJ)
PHATRDRAFT_37734 (Nop25 superfamily)
PHATRDRAFT_44564 (DUF4246)
PHATRDRAFT_44627 gtp-binding protein (Obg)
PHATRDRAFT_47281 pentatricopeptiderepeat-containing protein
PHATRDRAFT_47705 ribosome receptor
PHATRDRAFT_49439 cbf1 interacting corepressor cir (Cir_N)
PHATRDRAFT_49534 biopterin transport-related protein bt1 (BT1)
PHATRDRAFT_49540 (MFS)
PHATRDRAFT_49642 pentatrichopeptide repeatprotein (PPR_2)
PHATRDRAFT_49765 (Peptidase_C48 superfamily)
PHATRDRAFT_49919 splicing endonuclease positive effector sen1 (AAA_12 superfamily)
PHATRDRAFT_52346 mgc88910 protein (Glyco_hydro_47)
PHATRDRAFT_8821 atp-bindingsub-family b (mdr tap)member 10 (ABC_ATPase superfamily)
PHATRDRAFT_9078 photosystem ii 11 kd protein (PSII_Pbs27)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006817 | phosphate transport | 0.0120481 | 1 | Phatr_bicluster_0063 |
| GO:0006508 | proteolysis and peptidolysis | 0.053216 | 1 | Phatr_bicluster_0063 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.08657 | 1 | Phatr_bicluster_0063 |
| GO:0006457 | protein folding | 0.185684 | 1 | Phatr_bicluster_0063 |
| GO:0006412 | protein biosynthesis | 0.247414 | 1 | Phatr_bicluster_0063 |
| GO:0006118 | electron transport | 0.33492 | 1 | Phatr_bicluster_0063 |


Comments