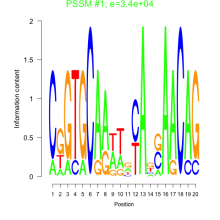
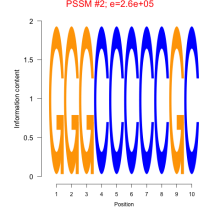
Phatr_bicluster_0066 Residual: 0.37
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0066 | 0.37 | Phaeodactylum tricornutum |
Displaying 1 - 30 of 30
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10747 flavin reductase-related (SDR_a5)
PHATRDRAFT_11792 psab translation factor (DUF1092 superfamily)
PHATRDRAFT_12097 zinc finger (c3hc4-type ring finger) family protein (zinc_ribbon_6)
PHATRDRAFT_12106 ku70-binding family protein (Peptidase_M76 superfamily)
PHATRDRAFT_2957 histone acetyltransferase (Bromodomain superfamily)
PHATRDRAFT_31822 nadph oxidoreductase (NADB_Rossmann superfamily)
PHATRDRAFT_34859 glyoxalase bleomycin resistance protein dioxygenase (Glo_EDI_BRP_like_6)
PHATRDRAFT_35733 (NADB_Rossmann superfamily)
PHATRDRAFT_36008 (DUF1632 superfamily)
PHATRDRAFT_36009 type i secretion target repeat
PHATRDRAFT_36269 type i secretion target repeat
PHATRDRAFT_41106 (PDZ superfamily)
PHATRDRAFT_43236 germination protein
PHATRDRAFT_45294 (TPR_12)
PHATRDRAFT_45346 endonuclease iii (ENDO3c)
PHATRDRAFT_45366 sepiapterin reductase (NADB_Rossmann superfamily)
PHATRDRAFT_45757 traf-type zinc finger family protein
PHATRDRAFT_46375 alkaline phosphatase d domain protein (MPP_PhoD)
PHATRDRAFT_46642 auxin efflux carrier family protein (Mem_trans superfamily)
PHATRDRAFT_8934 (DUF303 superfamily)
PHATRDRAFT_9664 cg14021-isoform b isoform 2
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0016567 | protein ubiquitination | 0.00618161 | 1 | Phatr_bicluster_0066 |
| GO:0006614 | SRP-dependent cotranslational protein-membrane targeting | 0.00861852 | 1 | Phatr_bicluster_0066 |
| GO:0006508 | proteolysis and peptidolysis | 0.053216 | 1 | Phatr_bicluster_0066 |
| GO:0006468 | protein amino acid phosphorylation | 0.262204 | 1 | Phatr_bicluster_0066 |
| GO:0008152 | metabolism | 0.410263 | 1 | Phatr_bicluster_0066 |

Comments