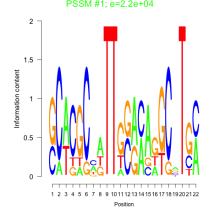
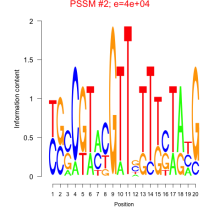
Phatr_bicluster_0069 Residual: 0.42
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0069 | 0.42 | Phaeodactylum tricornutum |
Displaying 1 - 22 of 22
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12969 haloacid dehalogenase-like hydrolase family protein (PLN02779)
PHATRDRAFT_29014 dhx33 protein (HA2)
PHATRDRAFT_32893 fructose-bisphosphate aldolase (FBP_aldolase_IIA)
PHATRDRAFT_39477 heavy metal translocating p-type atpase
PHATRDRAFT_43365 (AlgLyase superfamily)
PHATRDRAFT_44969 galactose-3-o-sulfotransferase 3
PHATRDRAFT_46399 abc1 familyprotein (AarF)
PHATRDRAFT_46770 amineflavin-containing superfamily (Amino_oxidase)
PHATRDRAFT_47408 y2104_arath uncharacterized methyltransferasechloroplast precursor (AdoMet_MTases)
PHATRDRAFT_50055 (ANK)
PHATRDRAFT_50634 ankyrin repeat (ANK)
PHATRDRAFT_7208 succinate dehydrogenase (PLN00129)
PHATRDRAFT_8171 cmp dcmpzinc-binding protein (nucleoside_deaminase)
PHATRDRAFT_8945 thioredoxin (TRX_family)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006096 | glycolysis | 0.0671528 | 1 | Phatr_bicluster_0069 |
| GO:0006118 | electron transport | 0.0711586 | 1 | Phatr_bicluster_0069 |
| GO:0008152 | metabolism | 0.109996 | 1 | Phatr_bicluster_0069 |
| GO:0006468 | protein amino acid phosphorylation | 0.2936 | 1 | Phatr_bicluster_0069 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.359328 | 1 | Phatr_bicluster_0069 |
| GO:0006508 | proteolysis and peptidolysis | 0.365994 | 1 | Phatr_bicluster_0069 |

Comments