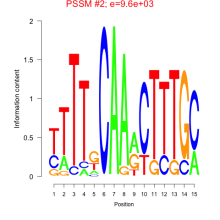
Phatr_bicluster_0073 Residual: 0.29
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0073 | 0.29 | Phaeodactylum tricornutum |
Displaying 1 - 33 of 33
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_18599 thioredoxin-like 4a (DIM1)
PHATRDRAFT_23292 l-galactono--lactone dehydrogenase (PLN02465)
PHATRDRAFT_25572 glutaryl-coenzyme a dehydrogenase (GCD)
PHATRDRAFT_29887 cg3714-isoform a (PLN02885)
PHATRDRAFT_33371 (DUF303 superfamily)
PHATRDRAFT_40783 hemolysin-related protein (TlyC)
PHATRDRAFT_44037 (Pep3_Vps18 superfamily)
PHATRDRAFT_46959 methyltransferase nucleic acid binding (COG1041)
PHATRDRAFT_47268 twin-arginine translocation protein e (TatA)
PHATRDRAFT_47271 polyprenyl synthetase (Trans_IPPS_HT)
PHATRDRAFT_47845 mucin-desulfating sulfatase (n-acetylglucosamine-6-sulfatase) (PRK13759)
PHATRDRAFT_47925 monooxygenase family protein (NAD_binding_8 superfamily)
PHATRDRAFT_48157 abscisic insensitive 1b (PP2Cc)
PHATRDRAFT_48449 chalcone-flavanone isomerase-related (Chalcone superfamily)
PHATRDRAFT_49014 (PH)
PHATRDRAFT_49435 (LRR_RI superfamily)
PHATRDRAFT_49552 purine nucleoside permease (Cupin_5 superfamily)
PHATRDRAFT_49707 endothelin-converting enzyme (PepO)
PHATRDRAFT_49782 guanine cytosine deaminase (CumB)
PHATRDRAFT_50212 ribosome recycling factor (RRF)
PHATRDRAFT_50229 pabp-dependent polynuclease 3 isoform 2 (PKc_like superfamily)
PHATRDRAFT_50511 (Bestrophin superfamily)
PHATRDRAFT_55040 d-lactate dehydrogenase (GlcD)
PHATRDRAFT_8113 blue light receptor (PAS_9)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019363 | pyridine nucleotide biosynthesis | 0.0103473 | 1 | Phatr_bicluster_0073 |
| GO:0030655 | beta-lactam antibiotic catabolism | 0.0154826 | 1 | Phatr_bicluster_0073 |
| GO:0007067 | mitosis | 0.0154826 | 1 | Phatr_bicluster_0073 |
| GO:0046677 | response to antibiotic | 0.0205926 | 1 | Phatr_bicluster_0073 |
| GO:0006118 | electron transport | 0.0278752 | 1 | Phatr_bicluster_0073 |
| GO:0008299 | isoprenoid biosynthesis | 0.0457657 | 1 | Phatr_bicluster_0073 |
| GO:0006811 | ion transport | 0.0556614 | 1 | Phatr_bicluster_0073 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0942755 | 1 | Phatr_bicluster_0073 |
| GO:0006725 | aromatic compound metabolism | 0.0989952 | 1 | Phatr_bicluster_0073 |
| GO:0006306 | DNA methylation | 0.0989952 | 1 | Phatr_bicluster_0073 |


Comments