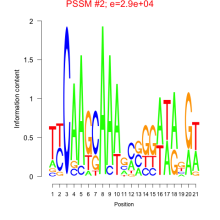
Phatr_bicluster_0078 Residual: 0.34
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0078 | 0.34 | Phaeodactylum tricornutum |
Displaying 1 - 19 of 19
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_16798 synaptobrevin 8-1 (Synaptobrevin)
PHATRDRAFT_27067 abc transporter like protein (AarF)
PHATRDRAFT_30461 sterol delta7 reductase (ERG4_ERG24 superfamily)
PHATRDRAFT_4074 atnadk-3 nadk3 (nadkinase 3) nad+ kinase nadh kinase (PLN02929)
PHATRDRAFT_42741 tpr repeat protein (Sulfotransfer_1 superfamily)
PHATRDRAFT_44044 npl4 family protein (MPN_NPL4)
PHATRDRAFT_44089 arp1 actin-related protein 1 homologcentractin alpha (ACTIN)
PHATRDRAFT_44716 proteinmembrane associated tyrosine threonine 1 (PKc)
PHATRDRAFT_45928 phytanoyl-dioxygenase (PhyH superfamily)
PHATRDRAFT_47187 2-polyprenyl-3-methyl-5-hydroxy-6-metoxy--benzoquinol methylase-like protein
PHATRDRAFT_48012 casein kinasedelta isoform 1 (Pkinase)
PHATRDRAFT_49335 duf647 family protein (DUF647)
PHATRDRAFT_49501 deacetylase sulfotransferase
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006468 | protein amino acid phosphorylation | 0.000707349 | 1 | Phatr_bicluster_0078 |
| GO:0016192 | vesicle-mediated transport | 0.0159584 | 1 | Phatr_bicluster_0078 |
| GO:0006810 | transport | 0.21473 | 1 | Phatr_bicluster_0078 |


Comments