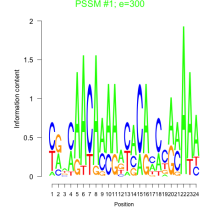
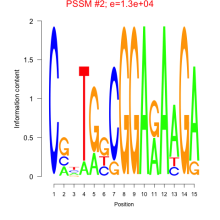
Phatr_bicluster_0083 Residual: 0.30
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0083 | 0.30 | Phaeodactylum tricornutum |
Displaying 1 - 33 of 33
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12312 atp-dependent rna helicase (HELICc)
PHATRDRAFT_13752 mitochondrial carrier protein (Mito_carr)
PHATRDRAFT_14138 trna (guanine-n-)-methyltransferase (Methyltransf_4 superfamily)
PHATRDRAFT_19122 dimethyladenosine transferase (AdoMet_MTases)
PHATRDRAFT_20342 nadh-glutamate synthase small chain (gltD)
PHATRDRAFT_3131 (Ras_like_GTPase superfamily)
PHATRDRAFT_32139 (Nop16 superfamily)
PHATRDRAFT_41091 (DUF2039 superfamily)
PHATRDRAFT_41785 eukaryotic initiation factor (PTZ00424)
PHATRDRAFT_42428 rna binding s1 (COG2996)
PHATRDRAFT_45079 diacylglycerol kinase (LCB5)
PHATRDRAFT_46371 methyltransfer with n-terminal ankyrin repeats (ANK)
PHATRDRAFT_46453 arystal structure of human choline kinase alpha 2 (APH_ChoK_like superfamily)
PHATRDRAFT_46999 deoxyhypusine synthase (DS)
PHATRDRAFT_47017 mms19l-prov protein (MMS19_N superfamily)
PHATRDRAFT_47242 (MFS_2)
PHATRDRAFT_47613 (Methyltransf_32 superfamily)
PHATRDRAFT_48912 c2h2 finger domain
PHATRDRAFT_49908 mus musculushomolog subfamily b member 5 (heat shock protein hsp40-3) (heat shock protein cognate 40) (DnaJ)
PHATRDRAFT_54092 myb domain-containing protein (SWIRM)
PHATRDRAFT_6934 myo-inositol-1-phosphate synthase family protein (NAD_binding_5 superfamily)
PHATRDRAFT_7634 dusp12 protein
PHATRDRAFT_9283 nadph-cytochrome p450 oxidoreductase (CysJ)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008612 | hypusine biosynthesis from peptidyl-lysine | 0.00296369 | 1 | Phatr_bicluster_0083 |
| GO:0050983 | spermidine catabolism to deoxyhypusine, using deoxyhypusine synthase | 0.00296369 | 1 | Phatr_bicluster_0083 |
| GO:0000154 | rRNA modification | 0.00591934 | 1 | Phatr_bicluster_0083 |
| GO:0007205 | protein kinase C activation | 0.00886694 | 1 | Phatr_bicluster_0083 |
| GO:0006537 | glutamate biosynthesis | 0.0147381 | 1 | Phatr_bicluster_0083 |
| GO:0006364 | rRNA processing | 0.0234852 | 1 | Phatr_bicluster_0083 |
| GO:0006118 | electron transport | 0.144823 | 1 | Phatr_bicluster_0083 |
| GO:0005975 | carbohydrate metabolism | 0.171749 | 1 | Phatr_bicluster_0083 |
| GO:0016567 | protein ubiquitination | 0.193944 | 1 | Phatr_bicluster_0083 |
| GO:0006457 | protein folding | 0.296964 | 1 | Phatr_bicluster_0083 |

Comments