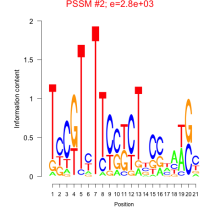
Phatr_bicluster_0095 Residual: 0.39
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0095 | 0.39 | Phaeodactylum tricornutum |
Displaying 1 - 31 of 31
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11230 like protein (BolA)
PHATRDRAFT_12927 (LRR_RI superfamily)
PHATRDRAFT_14599 rna polymerase sigma factor (Sig70-cyanoRpoD)
PHATRDRAFT_14867 uncharacterized conserved protein (Fe-S_biosyn superfamily)
PHATRDRAFT_1598 hippocampus abundant gene transcript 1 (MFS_1)
PHATRDRAFT_16076 at3g50680 t3a5_60
PHATRDRAFT_16334 at3g61870 f21f14_40 (DUF4079)
PHATRDRAFT_19030 mc family transporter: phosphate (Mito_carr)
PHATRDRAFT_2217 histidine decarboxylase (PLN03032)
PHATRDRAFT_37144 short-chain alcohol dehydrogenase-like protein (NADB_Rossmann superfamily)
PHATRDRAFT_37545 rna polymerase ii transcriptional (PC4)
PHATRDRAFT_41409 rna helicase - like protein (Dtyr_deacylase superfamily)
PHATRDRAFT_44383 (NADB_Rossmann superfamily)
PHATRDRAFT_44612 eif4g-related protein nat1 (W2_eIF4G1_like)
PHATRDRAFT_45205 ribonuclease h i (RNase_HI_like)
PHATRDRAFT_4604 heat shock factor (HSF_DNA-bind superfamily)
PHATRDRAFT_46218 af375428_1at4g29590 t16l4_100
PHATRDRAFT_47959 aldo keto reductase family protein (Aldo_ket_red)
PHATRDRAFT_48260 (Peptidase_M16 superfamily)
PHATRDRAFT_48861 erg24_schpo delta-sterol reductase (c-14 sterol reductase) (sterol c14-reductase) (ERG4_ERG24 superfamily)
PHATRDRAFT_54420 lumenal-like protein (PsbP superfamily)
PHATRDRAFT_54920 gtp binding protein (Sar1)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019478 | D-amino acid catabolism | 0.00370462 | 1 | Phatr_bicluster_0095 |
| GO:0015904 | tetracycline transport | 0.0147418 | 1 | Phatr_bicluster_0095 |
| GO:0006446 | regulation of translational initiation | 0.0147418 | 1 | Phatr_bicluster_0095 |
| GO:0006352 | transcription initiation | 0.0436188 | 1 | Phatr_bicluster_0095 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0438421 | 1 | Phatr_bicluster_0095 |
| GO:0006508 | proteolysis and peptidolysis | 0.0464053 | 1 | Phatr_bicluster_0095 |
| GO:0006520 | amino acid metabolism | 0.0854566 | 1 | Phatr_bicluster_0095 |
| GO:0006810 | transport | 0.155631 | 1 | Phatr_bicluster_0095 |
| GO:0008152 | metabolism | 0.298106 | 1 | Phatr_bicluster_0095 |

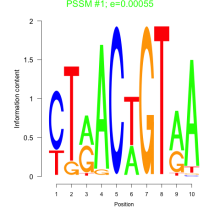
Comments