Phatr_bicluster_0097 Residual: 0.27
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0097 | 0.27 | Phaeodactylum tricornutum |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11441 rna polymerase ii second largest subunit (PRK08565)
PHATRDRAFT_12239 adenylate kinase (adk)
PHATRDRAFT_14995 cell division protein (FtsZ_type1)
PHATRDRAFT_15764 glutathione s-transferase (Gst)
PHATRDRAFT_20708 aspartyl aminopeptidase (M18_DAP)
PHATRDRAFT_22388 membrane protein (PRX5_like)
PHATRDRAFT_22459 delta-5 fatty acid desaturase (Delta6-FADS-like)
PHATRDRAFT_35819 glutathione reductase (PRK06116)
PHATRDRAFT_40952 (PUB)
PHATRDRAFT_41676 nucleoside diphosphate kinase family protein (NDPk_I)
PHATRDRAFT_42018 chloroplast ferredoxin nadp(+) reductase (PLN03116)
PHATRDRAFT_44172 acid phosphataselysophosphatidic (HP_HAP_like)
PHATRDRAFT_45692 nuclear exosome component rrp6p (DnaQ_like_exo superfamily)
PHATRDRAFT_47954 mitochondrial phosphate carrier protein (Mito_carr)
PHATRDRAFT_51100 lsm (like-sm) domain-containing protein (Sm_like superfamily)
PHATRDRAFT_54285 p-atpase family transporter: cation (P-ATPase-V)
PHATRDRAFT_6834 adenine phosphoribosyltransferase (PRTases_typeI)
PHATRDRAFT_7784 uncharacterized conserved membrane protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006749 | glutathione metabolism | 0.00419857 | 1 | Phatr_bicluster_0097 |
| GO:0006241 | CTP biosynthesis | 0.012546 | 1 | Phatr_bicluster_0097 |
| GO:0006183 | GTP biosynthesis | 0.012546 | 1 | Phatr_bicluster_0097 |
| GO:0006228 | UTP biosynthesis | 0.012546 | 1 | Phatr_bicluster_0097 |
| GO:0006118 | electron transport | 0.0132312 | 1 | Phatr_bicluster_0097 |
| GO:0051258 | protein polymerization | 0.0208275 | 1 | Phatr_bicluster_0097 |
| GO:0009116 | nucleoside metabolism | 0.0371948 | 1 | Phatr_bicluster_0097 |
| GO:0006636 | fatty acid desaturation | 0.0412462 | 1 | Phatr_bicluster_0097 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 0.0452816 | 1 | Phatr_bicluster_0097 |
| GO:0006350 | transcription | 0.0847655 | 1 | Phatr_bicluster_0097 |

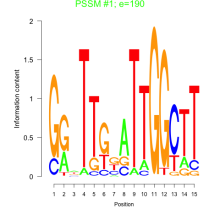

Comments