Phatr_bicluster_0099 Residual: 0.44
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0099 | 0.44 | Phaeodactylum tricornutum |
Displaying 1 - 56 of 56
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_13230 helicase domain protein (HA)
PHATRDRAFT_1813 ammonium transporter (Ammonium_transp superfamily)
PHATRDRAFT_33648 lipopolysaccharide a protein (CAP10 superfamily)
PHATRDRAFT_34415 heat shock transcription factor 4 isoform 2 (HSF_DNA-bind)
PHATRDRAFT_36581 transposonen spm sub-expressed
PHATRDRAFT_37892 (Bestrophin superfamily)
PHATRDRAFT_38600 tpr repeats
PHATRDRAFT_38934 adhesion lipoprotein (Fasciclin)
PHATRDRAFT_40447 (SMK-1 superfamily)
PHATRDRAFT_40510 protein disulfide isomerase-like protein 3 (Thioredoxin_like superfamily)
PHATRDRAFT_40749 (Glyco_tranf_GTA_type superfamily)
PHATRDRAFT_40874 exostosin family protein (Exostosin superfamily)
PHATRDRAFT_41350 probable cell surface glycoprotein
PHATRDRAFT_42685 hmg family member (hmg-4) (HMGB-UBF_HMG-box)
PHATRDRAFT_42955 ipt tig domain containing protein
PHATRDRAFT_43925 viral a-type inclusion (Smc)
PHATRDRAFT_44890 chitin synthase (Glyco_tranf_GTA_type superfamily)
PHATRDRAFT_45159 cog4325: membrane protein (DUF2254)
PHATRDRAFT_45771 secreted protein
PHATRDRAFT_49605 (Glyco_tranf_GTA_type superfamily)
PHATRDRAFT_49918 (NADB_Rossmann superfamily)
PHATRDRAFT_49986 m6 (immune inhibitor a) family (Peptidase_M6 superfamily)
PHATRDRAFT_54105 extracellular matrix protein st310
PHATRDRAFT_5465 (Glyoxalase_3)
PHATRDRAFT_944 ring finger and chy zinc finger domain containing 1 (zf-CHY)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007155 | cell adhesion | 0.0222757 | 1 | Phatr_bicluster_0099 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0510752 | 1 | Phatr_bicluster_0099 |
| GO:0016567 | protein ubiquitination | 0.118111 | 1 | Phatr_bicluster_0099 |
| GO:0006468 | protein amino acid phosphorylation | 0.262204 | 1 | Phatr_bicluster_0099 |
| GO:0006810 | transport | 0.287164 | 1 | Phatr_bicluster_0099 |
| GO:0006118 | electron transport | 0.33492 | 1 | Phatr_bicluster_0099 |

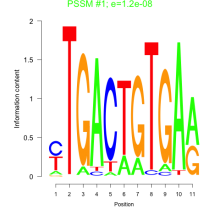

Comments