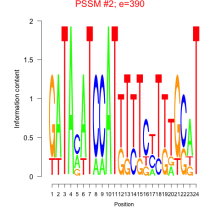
Phatr_bicluster_0105 Residual: 0.29
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0105 | 0.29 | Phaeodactylum tricornutum |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11014 acyl-dehydrogenase (DUF1974 superfamily)
PHATRDRAFT_12373 mgc114925 protein (ETF_alpha)
PHATRDRAFT_14457 mgc107891 protein (rpa1)
PHATRDRAFT_17427 alcohol dehydrogenase family protein (adh_III_F_hyde)
PHATRDRAFT_28068 mitochondrial trifunctionalbeta subunit (fadI)
PHATRDRAFT_32830 ribonucleaselarge subunit (RNase_HII_eukaryota_like)
PHATRDRAFT_35463 protein kinase p46g22 (S_TKc)
PHATRDRAFT_43398 glutathione reductase
PHATRDRAFT_43943 far-red impaired response protein
PHATRDRAFT_45811 (SEC14)
PHATRDRAFT_45947 acetyl-coenzyme a acyltransferase 2 (mitochondrial 3-oxoacyl-coenzyme a thiolase) (PRK05790)
PHATRDRAFT_46606 membrane protein
PHATRDRAFT_46793 cyst wall protein 1
PHATRDRAFT_48922 loc398088 protein (Cnd3)
PHATRDRAFT_53929 mevalonate kinase (ERG12)
PHATRDRAFT_54306 microtubule-associated protein eb1-like protein (EB1)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0016070 | RNA metabolism | 0.00123487 | 1 | Phatr_bicluster_0105 |
| GO:0006118 | electron transport | 0.0284292 | 1 | Phatr_bicluster_0105 |
| GO:0006260 | DNA replication | 0.0317125 | 1 | Phatr_bicluster_0105 |
| GO:0006468 | protein amino acid phosphorylation | 0.195192 | 1 | Phatr_bicluster_0105 |


Comments