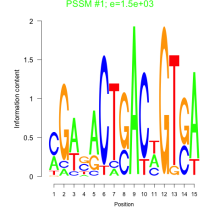
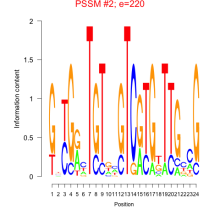
Phatr_bicluster_0107 Residual: 0.29
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0107 | 0.29 | Phaeodactylum tricornutum |
Displaying 1 - 28 of 28
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10830 ferredoxin-dependent bilin reductase (Fe_bilin_red superfamily)
PHATRDRAFT_12420 amp-dependent synthetase and ligase (FAA1)
PHATRDRAFT_16545 methionyl-trna synthetase (tRNA_bind_EMAP-II_like)
PHATRDRAFT_18893 quinone oxidoreductase (QOR2)
PHATRDRAFT_23639 usp_cucmeudp-sugar pyrophospharylase (udp-galactose glucose pyrophosphorylase) (PLN02830)
PHATRDRAFT_29157 phosphoglycerate kinase (Phosphoglycerate_kinase)
PHATRDRAFT_30113 lipoamide dehydrogenase (PRK06416)
PHATRDRAFT_31409 thioesterase family protein (HopJ superfamily)
PHATRDRAFT_33530 mpbq msbq transferase cyanobacterial type (AdoMet_MTases)
PHATRDRAFT_33543 hydrogenase component
PHATRDRAFT_37652 malonyl-:acp transacylase (PLN02752)
PHATRDRAFT_41856 plastid transketolase (PLN02790)
PHATRDRAFT_41878 phytoene synthase (Trans_IPPS_HH)
PHATRDRAFT_43169 mrna binding protein precursor (PLN00016)
PHATRDRAFT_43831 at1g26760 t24p13_13 (GH43_62_32_68 superfamily)
PHATRDRAFT_44335 at1g06690 f4h5_17 (Aldo_ket_red superfamily)
PHATRDRAFT_44486 peptidase m50 (S2P-M50_like_2)
PHATRDRAFT_44651 dimethylmenaquinone methyltransferase (Methyltransf_6 superfamily)
PHATRDRAFT_45846 violaxanthin de-epoxidasechloroplast (VDE superfamily)
PHATRDRAFT_46395 (DUF1118 superfamily)
PHATRDRAFT_51703 violaxanthin de-epoxidase (VDE superfamily)
PHATRDRAFT_53935 ribulose-phosphate 3-epimerase (RPE)
PHATRDRAFT_54082 malic enzyme (PLN03129)
PHATRDRAFT_9312 sigma-70 factor (Sig70-cyanoRpoD)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0010024 | phytochromobilin biosynthesis | 0.00886694 | 1 | Phatr_bicluster_0107 |
| GO:0006352 | transcription initiation | 0.0350372 | 1 | Phatr_bicluster_0107 |
| GO:0006633 | fatty acid biosynthesis | 0.0379055 | 1 | Phatr_bicluster_0107 |
| GO:0006306 | DNA methylation | 0.0577662 | 1 | Phatr_bicluster_0107 |
| GO:0006096 | glycolysis | 0.0990654 | 1 | Phatr_bicluster_0107 |
| GO:0009058 | biosynthesis | 0.125669 | 1 | Phatr_bicluster_0107 |
| GO:0006508 | proteolysis and peptidolysis | 0.13969 | 1 | Phatr_bicluster_0107 |
| GO:0008152 | metabolism | 0.21493 | 1 | Phatr_bicluster_0107 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.487367 | 1 | Phatr_bicluster_0107 |
| GO:0006118 | electron transport | 0.503223 | 1 | Phatr_bicluster_0107 |

Comments