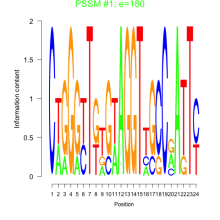
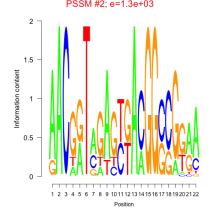
Phatr_bicluster_0112 Residual: 0.35
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0112 | 0.35 | Phaeodactylum tricornutum |
Displaying 1 - 28 of 28
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_16401 endoplasmic reticulum nucleotide sugar transporter (UAA)
PHATRDRAFT_19669 vacuolar protein sorting 28 (VPS28 superfamily)
PHATRDRAFT_32087 cyclin a homolog (Cyclin_C)
PHATRDRAFT_37509 cytochrome c553 (Cytochrom_C superfamily)
PHATRDRAFT_37918 death-specific protein (EFh)
PHATRDRAFT_40147 novel protein (ANK)
PHATRDRAFT_43891 cct motif family protein (CCT)
PHATRDRAFT_48112 dihydroorotate dehydrogenase (TIM_phosphate_binding superfamily)
PHATRDRAFT_48337 kinesin light chain (TPR_12)
PHATRDRAFT_49632 loc507659 protein (UEV superfamily)
PHATRDRAFT_49665 ser thr protein phosphatase family (MPP_superfamily superfamily)
PHATRDRAFT_50366 viral a-type inclusion
PHATRDRAFT_50516 probable na+ h+ antiporter (Na_H_Exchanger)
PHATRDRAFT_7736 synaptic vesicle transporter svop and related transporters (major facilitator superfamily) (COG5134 superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006207 | 'de novo' pyrimidine base biosynthesis | 0.00493217 | 1 | Phatr_bicluster_0112 |
| GO:0015031 | protein transport | 0.0377177 | 1 | Phatr_bicluster_0112 |
| GO:0000074 | regulation of cell cycle | 0.0413065 | 1 | Phatr_bicluster_0112 |
| GO:0006512 | ubiquitin cycle | 0.0460749 | 1 | Phatr_bicluster_0112 |
| GO:0006464 | protein modification | 0.0754495 | 1 | Phatr_bicluster_0112 |

Comments