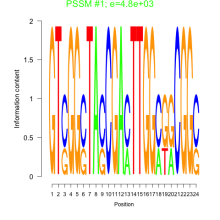
Phatr_bicluster_0120 Residual: 0.36
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0120 | 0.36 | Phaeodactylum tricornutum |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10593 atp-dependent clp protease proteolytic subunit (S14_ClpP_2)
PHATRDRAFT_13921 microsomal signal peptidase 12 kda subunit family protein (SPC12)
PHATRDRAFT_14472 flj43980 protein (ANK)
PHATRDRAFT_14762 isocitrate dehydrogenase (Iso_dh superfamily)
PHATRDRAFT_14927 frda_schpo frataxinmitochondrial precursor (Frataxin superfamily)
PHATRDRAFT_15214 at1g65820 f1e22_4
PHATRDRAFT_18665 shm4 (serine hydroxymethyltransferase 4) glycine hydroxymethyltransferase (SHMT)
PHATRDRAFT_19093 mgc80063 protein (Coatomer_E)
PHATRDRAFT_21535 af228419_1rab6 protein (Rab6)
PHATRDRAFT_22896 (ArsA_ATPase)
PHATRDRAFT_29758 puromycin sensitive aminopeptidase cg1009-isoform a (M1_APN_2)
PHATRDRAFT_30471 methylenetetrahydrofolate reductase (MTHFR)
PHATRDRAFT_37658 glutathione s-transferase-like protein (GST_N_Sigma_like)
PHATRDRAFT_40933 atp-dependent clpproteolytic subunit (S14_ClpP_2)
PHATRDRAFT_43061 dna j-like protein 1 (DnaJ)
PHATRDRAFT_50476 glutathione peroxidase-like protein (Thioredoxin_like superfamily)
PHATRDRAFT_51242 adenosine kinase isoform 1t-like protein (PTZ00247)
PHATRDRAFT_54589 sec61beta family protein
PHATRDRAFT_6756 (HAM1)
PHATRDRAFT_7237 transport protein (Sm_multidrug_ex)
PHATRDRAFT_7528 nucleoredoxin 1 (Thioredoxin_like superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006166 | purine ribonucleoside salvage | 0.00370462 | 1 | Phatr_bicluster_0120 |
| GO:0006563 | L-serine metabolism | 0.00739642 | 1 | Phatr_bicluster_0120 |
| GO:0006555 | methionine metabolism | 0.00739642 | 1 | Phatr_bicluster_0120 |
| GO:0006886 | intracellular protein transport | 0.00921303 | 1 | Phatr_bicluster_0120 |
| GO:0006544 | glycine metabolism | 0.0110755 | 1 | Phatr_bicluster_0120 |
| GO:0006465 | signal peptide processing | 0.0147418 | 1 | Phatr_bicluster_0120 |
| GO:0006820 | anion transport | 0.0436188 | 1 | Phatr_bicluster_0120 |
| GO:0006508 | proteolysis and peptidolysis | 0.0464053 | 1 | Phatr_bicluster_0120 |
| GO:0007264 | small GTPase mediated signal transduction | 0.0751604 | 1 | Phatr_bicluster_0120 |
| GO:0015031 | protein transport | 0.109067 | 1 | Phatr_bicluster_0120 |

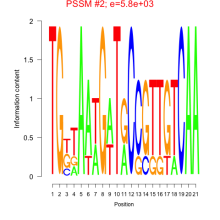
Comments