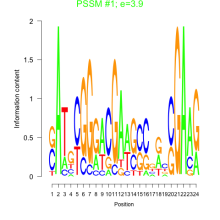
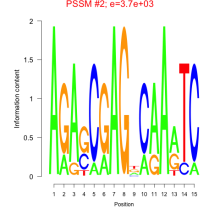
Phatr_bicluster_0125 Residual: 0.29
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0125 | 0.29 | Phaeodactylum tricornutum |
Displaying 1 - 35 of 35
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11735 26s proteasome aaa-atpase subunit rpt3 (PTZ00454)
PHATRDRAFT_13622 mgc83233 protein (RPN1)
PHATRDRAFT_19004 proteasome subunit alpha type 7 (proteasome_alpha_type_7)
PHATRDRAFT_19300 loc446265 protein (RPN2)
PHATRDRAFT_19341 proteasome chain protein (proteasome_beta_type_2)
PHATRDRAFT_20227 26s proteasome regulatory complex subunit rpn11 (MPN_RPN11_CSN5)
PHATRDRAFT_20434 at1g64520 f1n19_10 (SAC3_GANP)
PHATRDRAFT_24865 proteasome 26s non-atpase subunit 12 (PCI superfamily)
PHATRDRAFT_28202 20s proteasome alpha subunit e (proteasome_alpha_type_5)
PHATRDRAFT_28219 26s proteasome atpase subunit (RPT1)
PHATRDRAFT_30003 proteasomebeta type 3 (proteasome_beta_type_3)
PHATRDRAFT_30067 26s proteasome regulatory particle triple-a atpase subunit6 (RPT1)
PHATRDRAFT_33525 heat shock protein 33 (HSP33)
PHATRDRAFT_41172 calr_chlrecalreticulin precursor (Calreticulin)
PHATRDRAFT_41778 26s proteasome subunit 4 (AAA)
PHATRDRAFT_42566 protein disulfide-isomerase-like proteinep2 precursor (PDI_a_family)
PHATRDRAFT_42757 p23 co-chaperone of hsp90 system (p23_hB-ind1_like)
PHATRDRAFT_45122 26s proteasome regulatory complex atpase rpt4 (RPT1)
PHATRDRAFT_47062 armadillo beta-catenin repeat family protein
PHATRDRAFT_47306 protein disulfide isomerase (PDI_a_family)
PHATRDRAFT_47887 proteasome activator subunit 1 (PA28_beta superfamily)
PHATRDRAFT_47905 nucleic acid binding protein (CSP_CDS)
PHATRDRAFT_47918 regulatory protein suaprga1 (MAM33 superfamily)
PHATRDRAFT_49172 (zf-Tim10_DDP)
PHATRDRAFT_49897 20s proteasome alpha subunit (proteasome_alpha_type_2)
PHATRDRAFT_51280 signal peptidase i (S26_SPase_I)
PHATRDRAFT_51691 beta 1 subunit of 20s proteasome (proteasome_beta_type_6)
PHATRDRAFT_52198 (RPN6)
PHATRDRAFT_5311 proteasome subunit beta 7 (proteasome_beta_type_7)
PHATRDRAFT_54971 (Smc)
PHATRDRAFT_55079 pyruvate kinase (PRK05826)
PHATRDRAFT_55215 heat shock (PRK05218)
PHATRDRAFT_5685 pag1 (20s proteasome alpha subunit g1) peptidase (Ntn_hydrolase superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0030163 | protein catabolism | 0.00000000488 | 0.0000113 | Phatr_bicluster_0125 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.0000000128 | 0.0000297 | Phatr_bicluster_0125 |
| GO:0045039 | mitochondrial inner membrane protein import | 0.0206078 | 1 | Phatr_bicluster_0125 |
| GO:0006626 | protein-mitochondrial targeting | 0.0206078 | 1 | Phatr_bicluster_0125 |
| GO:0000074 | regulation of cell cycle | 0.0225725 | 1 | Phatr_bicluster_0125 |
| GO:0006465 | signal peptide processing | 0.0273856 | 1 | Phatr_bicluster_0125 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 0.0735796 | 1 | Phatr_bicluster_0125 |
| GO:0006260 | DNA replication | 0.165551 | 1 | Phatr_bicluster_0125 |
| GO:0006457 | protein folding | 0.193151 | 1 | Phatr_bicluster_0125 |
| GO:0006281 | DNA repair | 0.210935 | 1 | Phatr_bicluster_0125 |

Comments