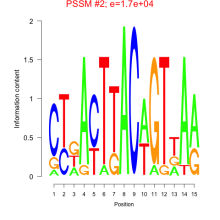
Phatr_bicluster_0126 Residual: 0.29
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0126 | 0.29 | Phaeodactylum tricornutum |
Displaying 1 - 26 of 26
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10367 non-transporter abc protein (Uup)
PHATRDRAFT_14618 atph+mitochondrial f1alpha subunitcardiac muscle (PRK09281)
PHATRDRAFT_14756 peptidase a22b family protein (Peptidase_A22B superfamily)
PHATRDRAFT_16376 fatty-acyl elongase (ELO)
PHATRDRAFT_18745 udp-glucose 6-expressed (PLN02353)
PHATRDRAFT_22774 ribosomal proteinisoform cra_a (L7)
PHATRDRAFT_22797 duf850 domain protein (DUF106)
PHATRDRAFT_23059 aspartatemitochondrial precursor (Aminotran_1_2)
PHATRDRAFT_23079 rl8_schpo 60s ribosomal protein l8 (Ribosomal_L7Ae superfamily)
PHATRDRAFT_31440 acyl carrier (PP-binding superfamily)
PHATRDRAFT_39687 ribosomal protein l22 (Ribosomal_L22e)
PHATRDRAFT_40052 polysaccharide deacetylase family protein (uraD_N-term-dom)
PHATRDRAFT_40492 dmt family transporter: phosphate phosphoenolpyruvate
PHATRDRAFT_43824 diphosphomevalonate decarboxylase (mevalonate pyrophosphate decarboxylase)
PHATRDRAFT_46969 cytoplasmicintermediate chain 2 (WD40 superfamily)
PHATRDRAFT_47104 (A_NRPS)
PHATRDRAFT_47675 rcc1-like g exchanging factor rlg
PHATRDRAFT_47749 chaperone protein (DnaJ)
PHATRDRAFT_48411 ribosomal protein s2 (rpsE_arch)
PHATRDRAFT_48457 transcriptional regulator (DUF179 superfamily)
PHATRDRAFT_49114 chaperonin containingsubunit 2 (TCP1_beta)
PHATRDRAFT_49608 assembly protein (UPF0051)
PHATRDRAFT_51026 histone deacetylase 1 (PTZ00063)
PHATRDRAFT_51169 gtp-binding nuclear protein ran (PTZ00132)
PHATRDRAFT_54805 cation-transporting atpase (MgtA)
PHATRDRAFT_6847 40s ribosomal proteinexpressed (PTZ00155)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006412 | protein biosynthesis | 0.00113258 | 1 | Phatr_bicluster_0126 |
| GO:0016575 | histone deacetylation | 0.0103473 | 1 | Phatr_bicluster_0126 |
| GO:0006754 | ATP biosynthesis | 0.0154826 | 1 | Phatr_bicluster_0126 |
| GO:0042254 | ribosome biogenesis and assembly | 0.0307368 | 1 | Phatr_bicluster_0126 |
| GO:0044267 | cellular protein metabolism | 0.0507259 | 1 | Phatr_bicluster_0126 |
| GO:0006633 | fatty acid biosynthesis | 0.0654593 | 1 | Phatr_bicluster_0126 |
| GO:0007264 | small GTPase mediated signal transduction | 0.103691 | 1 | Phatr_bicluster_0126 |
| GO:0006520 | amino acid metabolism | 0.117641 | 1 | Phatr_bicluster_0126 |
| GO:0015031 | protein transport | 0.149389 | 1 | Phatr_bicluster_0126 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.175733 | 1 | Phatr_bicluster_0126 |

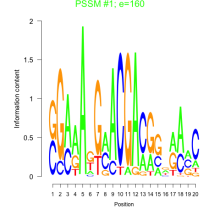
Comments