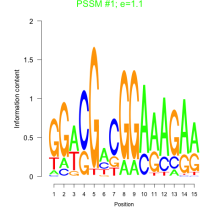
Phatr_bicluster_0127 Residual: 0.23
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0127 | 0.23 | Phaeodactylum tricornutum |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10251 (HELICc)
PHATRDRAFT_1241 dna topoisomerase i (TOP1Ac)
PHATRDRAFT_14149 rrp8_schpo ribosomal rna-processing protein 8 (AdoMet_MTases superfamily)
PHATRDRAFT_15233 5-3 exoribonuclease 2 (XRN_N superfamily)
PHATRDRAFT_18524 threonyl-trna synthetase (PLN02908)
PHATRDRAFT_23838 (COG0714)
PHATRDRAFT_24195 (CPSaseII_lrg)
PHATRDRAFT_33217 riken cdna 2410002o22isoform cra_a (DUF974 superfamily)
PHATRDRAFT_3351 dimethyladenosine transferase
PHATRDRAFT_3425 trna (5-methylaminomethyl-2-thiouridylate)-methyltransferase (tRNA_Me_trans)
PHATRDRAFT_38891 histidinol-phosphate aminotransferase (PLN03026)
PHATRDRAFT_4048 cg3645-isoform a (DUS_like_FMN)
PHATRDRAFT_4049 cg3645-isoform a (DUS_like_FMN)
PHATRDRAFT_41071 methyltransferase type 12 (AdoMet_MTases)
PHATRDRAFT_44042 exonuclease family protein (DnaQ_like_exo superfamily)
PHATRDRAFT_50465 (Smc)
PHATRDRAFT_50527 nucleolar gtpase atpase p130 (SRP40_C superfamily)
PHATRDRAFT_5175 solute carrier family 25 (mitochondrial carrier ornithine transporter) member 15 (Mito_carr)
PHATRDRAFT_7810 dead deah box helicase domain protein (SrmB)
PHATRDRAFT_8000 rnafamily protein (SpoU_methylase superfamily)
PHATRDRAFT_8457 (RRM_SF superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008033 | tRNA processing | 0.000171452 | 0.393825 | Phatr_bicluster_0127 |
| GO:0051341 | regulation of oxidoreductase activity | 0.00352946 | 1 | Phatr_bicluster_0127 |
| GO:0006435 | threonyl-tRNA aminoacylation | 0.01133 | 1 | Phatr_bicluster_0127 |
| GO:0000154 | rRNA modification | 0.01133 | 1 | Phatr_bicluster_0127 |
| GO:0019856 | pyrimidine base biosynthesis | 0.0169488 | 1 | Phatr_bicluster_0127 |
| GO:0006304 | DNA modification | 0.028095 | 1 | Phatr_bicluster_0127 |
| GO:0006268 | DNA unwinding | 0.028095 | 1 | Phatr_bicluster_0127 |
| GO:0006526 | arginine biosynthesis | 0.0391202 | 1 | Phatr_bicluster_0127 |
| GO:0006364 | rRNA processing | 0.0445878 | 1 | Phatr_bicluster_0127 |
| GO:0000105 | histidine biosynthesis | 0.0500257 | 1 | Phatr_bicluster_0127 |

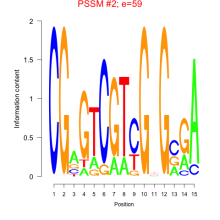
Comments