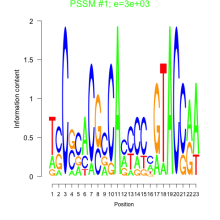
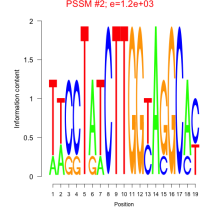
Phatr_bicluster_0135 Residual: 0.29
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0135 | 0.29 | Phaeodactylum tricornutum |
Displaying 1 - 36 of 36
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_14143 transcription factor cmyb isoform 1 (SANT)
PHATRDRAFT_16408 lysosomal trafficking regulator lyst and related beach and wd40 repeat proteins (Beach)
PHATRDRAFT_16608 rufy1 isoform 4 (FYVE)
PHATRDRAFT_16649 hydroxymethylglutaryl coenzyme a synthase (PLN02577)
PHATRDRAFT_16891 adapter-related protein complex 4 epsilon 1 (Adaptin_N)
PHATRDRAFT_29736 urate oxidase (TFold superfamily)
PHATRDRAFT_33571 heat shock protein 83-1
PHATRDRAFT_37460 cyclin serine peptidase familypeptidase (GAT_1 superfamily)
PHATRDRAFT_40578 ubiquitin-protein ligase e3-alpha-like protein (zf-UBR)
PHATRDRAFT_40677 (PH_BEACH)
PHATRDRAFT_41011 hect e3 ubiquitin ligase (RCC1)
PHATRDRAFT_41214 (Apc1 superfamily)
PHATRDRAFT_43898 (Sgf11)
PHATRDRAFT_44803 conserved protein (fungal and protazoan) (DUF2009 superfamily)
PHATRDRAFT_45008 camk family protein kinase (PKc_like superfamily)
PHATRDRAFT_47516 c3hc4 zinc-binding integral peroxisomal membrane protein (RING)
PHATRDRAFT_49706 (E_set_GO_C)
PHATRDRAFT_49954 core 1 udp-galactose:n-acetylgalactosamine-alpha-r beta- galactosy
PHATRDRAFT_54772 cop9 complex homolog subunit 1 b cg3889-pa (RPN7 superfamily)
PHATRDRAFT_55114 gap related family member (rbg-3) (TBC)
PHATRDRAFT_7866 dna mismatch repair protein muts (ABC_ATPase superfamily)
PHATRDRAFT_7867 s-adenosylmethionine decarboxylase related (AdoMet_dc)
PHATRDRAFT_8141 dna damage response protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006084 | acetyl-CoA metabolism | 0.00271672 | 1 | Phatr_bicluster_0135 |
| GO:0006144 | purine base metabolism | 0.00271672 | 1 | Phatr_bicluster_0135 |
| GO:0006298 | mismatch repair | 0.0162 | 1 | Phatr_bicluster_0135 |
| GO:0006512 | ubiquitin cycle | 0.0986411 | 1 | Phatr_bicluster_0135 |
| GO:0006508 | proteolysis and peptidolysis | 0.120633 | 1 | Phatr_bicluster_0135 |
| GO:0016567 | protein ubiquitination | 0.17931 | 1 | Phatr_bicluster_0135 |
| GO:0008152 | metabolism | 0.187614 | 1 | Phatr_bicluster_0135 |
| GO:0006457 | protein folding | 0.275986 | 1 | Phatr_bicluster_0135 |
| GO:0006468 | protein amino acid phosphorylation | 0.380037 | 1 | Phatr_bicluster_0135 |

Comments