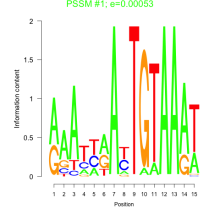
Phatr_bicluster_0136 Residual: 0.58
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0136 | 0.58 | Phaeodactylum tricornutum |
Displaying 1 - 35 of 35
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_18481 isoamyl acetate-hydrolyzing esterase 1 homolog (Isoamyl_acetate_hydrolase_like)
PHATRDRAFT_33400 (dsDNA_bind superfamily)
PHATRDRAFT_38108 receptor protein kinase (PLN00113)
PHATRDRAFT_39914 homocysteine methyltransferase (S-methyl_trans superfamily)
PHATRDRAFT_40462 (Tryp_SPc)
PHATRDRAFT_40785 core 1 udp-galactose:n-acetylgalactosamine-alpha-r beta- galactosy
PHATRDRAFT_44188 protein kinase domain containing protein (Methyltransf_31)
PHATRDRAFT_4423 histone deactylase of possible bacterial origin with ankyrin repeats at the n-terminus (HDAC_classII_2)
PHATRDRAFT_47565 amp-dependent synthetase and ligase (AFD_class_I superfamily)
PHATRDRAFT_49107 lipopolysaccharide a protein (CAP10 superfamily)
PHATRDRAFT_49511 (TPD superfamily)
PHATRDRAFT_49745 helicase domain protein (HA)
PHATRDRAFT_50357 probable cell surface glycoprotein
PHATRDRAFT_9609 lipophosphoglycan biosynthetic protein (VRG4)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006508 | proteolysis and peptidolysis | 0.00880858 | 1 | Phatr_bicluster_0136 |
| GO:0005975 | carbohydrate metabolism | 0.0459669 | 1 | Phatr_bicluster_0136 |

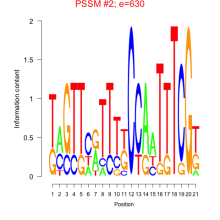
Comments