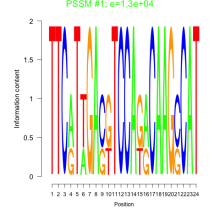
Phatr_bicluster_0141 Residual: 0.51
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0141 | 0.51 | Phaeodactylum tricornutum |
Displaying 1 - 21 of 21
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_26405 zinc transporter zip (Zip superfamily)
PHATRDRAFT_33729 cellulosomal scaffoldin anchoring protein (DDE_Tnp_1_7 superfamily)
PHATRDRAFT_34132 (DUF3291)
PHATRDRAFT_34626 high mobility group protein 4 (hmg-4) (high mobility group protein 2a) (hmg-2a) (HMGB-UBF_HMG-box)
PHATRDRAFT_35442 piggybac transposase uribo2 (DDE_Tnp_1_7 superfamily)
PHATRDRAFT_35939 short-chain dehydrogenase (fabG)
PHATRDRAFT_37297 cellulosomal scaffoldin anchoring protein (DDE_Tnp_1_7 superfamily)
PHATRDRAFT_38409 retroelement pol polyprotein
PHATRDRAFT_46486 large protein (Glyco_transf_49 superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0030001 | metal ion transport | 0.0118109 | 1 | Phatr_bicluster_0141 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.15368 | 1 | Phatr_bicluster_0141 |
| GO:0008152 | metabolism | 0.202444 | 1 | Phatr_bicluster_0141 |


Comments