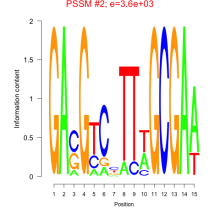
Phatr_bicluster_0142 Residual: 0.29
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0142 | 0.29 | Phaeodactylum tricornutum |
Displaying 1 - 26 of 26
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_21961 serine threonine-protein kinase ctr1 (S_TKc)
PHATRDRAFT_34078 ubiquinone menaquinone biosynthesis-related protein (AdoMet_MTases)
PHATRDRAFT_34485 fatty acid elongase (ELO)
PHATRDRAFT_35519 riken cdna 4832428d23 isoform 1 (Methyltransf_16 superfamily)
PHATRDRAFT_39978 amino acid transporter (Aa_trans superfamily)
PHATRDRAFT_40950 (DUF1989)
PHATRDRAFT_41055 (DUF914)
PHATRDRAFT_41356 (DUF2061 superfamily)
PHATRDRAFT_44207 (Anticodon_Ia_like superfamily)
PHATRDRAFT_45969 aaap family transporter: amino acid (Aa_trans superfamily)
PHATRDRAFT_47763 (DUF1295 superfamily)
PHATRDRAFT_48186 indole-3-glycerol-phosphate synthase (TIM_phosphate_binding superfamily)
PHATRDRAFT_48206 exonuclease ib (PIN_EXO1)
PHATRDRAFT_48502 trf4 5 nucletotidyl transferase
PHATRDRAFT_48781 kinesin light chain isoform 1
PHATRDRAFT_49621 (DnaQ_like_exo superfamily)
PHATRDRAFT_50482 (Arginase_HDAC superfamily)
PHATRDRAFT_5396 u7 family protein (S49_Sppa_N_C)
PHATRDRAFT_6093 set domain (SET)
PHATRDRAFT_7088 camp-dependent protein kinase type 3 (PKc_like superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006865 | amino acid transport | 0.00125869 | 1 | Phatr_bicluster_0142 |
| GO:0006568 | tryptophan metabolism | 0.0118036 | 1 | Phatr_bicluster_0142 |
| GO:0006468 | protein amino acid phosphorylation | 0.0424353 | 1 | Phatr_bicluster_0142 |
| GO:0006281 | DNA repair | 0.0652904 | 1 | Phatr_bicluster_0142 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.359328 | 1 | Phatr_bicluster_0142 |
| GO:0006508 | proteolysis and peptidolysis | 0.365994 | 1 | Phatr_bicluster_0142 |

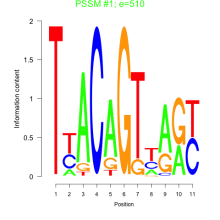
Comments