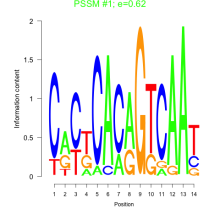
Phatr_bicluster_0144 Residual: 0.19
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0144 | 0.19 | Phaeodactylum tricornutum |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11311 silicon transporter (Silic_transp superfamily)
PHATRDRAFT_14639 at5g36790 f5h8_20 (HAD-SF-IIA)
PHATRDRAFT_21410 mitogen-activated protein kinase (STKc_MAPK)
PHATRDRAFT_32938 potassiumsubfamilymember 16 (Ion_trans_2)
PHATRDRAFT_42677 (CRCB)
PHATRDRAFT_42743 at4g27720 t29a15_210 (SAP)
PHATRDRAFT_43046 membrane protein (Rhomboid)
PHATRDRAFT_43667 cyclin b3 (CYCLIN)
PHATRDRAFT_43775 glyoxalase hydroxyacylglutathione (Lactamase_B superfamily)
PHATRDRAFT_44905 gamma-tubulin interacting protein (Spc97_Spc98)
PHATRDRAFT_45254 dolichyl glycosyltransferase (Alg6_Alg8 superfamily)
PHATRDRAFT_47846 viral a-type inclusion
PHATRDRAFT_49355 pheromone-regulated membrane protein prm10 (COG2966)
PHATRDRAFT_50345 floral homeotic protein hua1 (zf-CCCH)
PHATRDRAFT_8598 floral homeotic protein hua1 (zf-CCCH)
PHATRDRAFT_8678 b-type cyclin (CYCLIN)
PHATRDRAFT_9185 loc495505 protein (vWFA superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008152 | metabolism | 0.453158 | 1 | Phatr_bicluster_0144 |
| GO:0000226 | microtubule cytoskeleton organization and biogenesis | 0.00394818 | 1 | Phatr_bicluster_0144 |
| GO:0006813 | potassium ion transport | 0.0388725 | 1 | Phatr_bicluster_0144 |
| GO:0006468 | protein amino acid phosphorylation | 0.2936 | 1 | Phatr_bicluster_0144 |
| GO:0006412 | protein biosynthesis | 0.277391 | 1 | Phatr_bicluster_0144 |
| GO:0000074 | regulation of cell cycle | 0.00185698 | 1 | Phatr_bicluster_0144 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.0983212 | 1 | Phatr_bicluster_0144 |

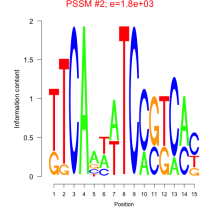
Comments