Phatr_bicluster_0150 Residual: 0.31
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0150 | 0.31 | Phaeodactylum tricornutum |
Displaying 1 - 26 of 26
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11308 sjchgc05278 protein (RRM_PPIE)
PHATRDRAFT_31375 chromatin modifying protein 1b (Snf7 superfamily)
PHATRDRAFT_37667 conserved protein (NADB_Rossmann superfamily)
PHATRDRAFT_38474 rna-binding region rnp-1 (RRM_SF)
PHATRDRAFT_38720 fucoxanthin chlorophyll a c binding protein (Chloroa_b-bind)
PHATRDRAFT_38985 coenzyme q biosynthesis protein (Coq4 superfamily)
PHATRDRAFT_42578 (PPI_Ypi1)
PHATRDRAFT_44043 loc495158 protein (Methyltransf_16 superfamily)
PHATRDRAFT_47847 (Arrestin_N superfamily)
PHATRDRAFT_48086 (META superfamily)
PHATRDRAFT_48482 ac007202_7 ests gb (DUF2358 superfamily)
PHATRDRAFT_48572 hsf-type dna-binding domain containing protein (HSF_DNA-bind superfamily)
PHATRDRAFT_54569 (BglC)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009765 | photosynthesis light harvesting | 0.0191736 | 1 | Phatr_bicluster_0150 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.105262 | 1 | Phatr_bicluster_0150 |

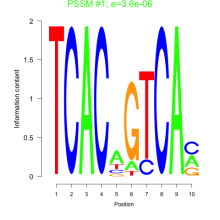
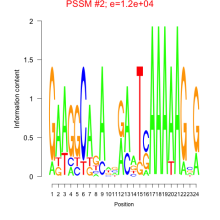
Comments