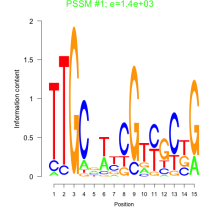
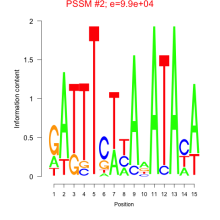
Phatr_bicluster_0153 Residual: 0.26
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0153 | 0.26 | Phaeodactylum tricornutum |
Displaying 1 - 19 of 19
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_23399 (metH)
PHATRDRAFT_28841 hypothetical maltose o-acetyltransferase (LbH_MAT_GAT)
PHATRDRAFT_29812 actin a (ACTIN)
PHATRDRAFT_31068 atp binding (ATP_bind_1)
PHATRDRAFT_43152 peptidyl-prolyl cis-transcyclophilin-type (cyclophilin_ABH_like)
PHATRDRAFT_48279 pentatricopeptide repeat protein
PHATRDRAFT_48744 glycosylfamily 2 (Glyco_tranf_GTA_type superfamily)
PHATRDRAFT_51136 (PRK12999)
PHATRDRAFT_51157 phosphoenolpyruvate carboxylase (PEPcase)
PHATRDRAFT_54935 actin a (ACTIN)
PHATRDRAFT_55192 clathrin coat assembly (APS2)
PHATRDRAFT_55198 enoyl-hydratase isomerase (PRK05862)
PHATRDRAFT_9904 oligosaccharyl transferase subunit (PMT_2 superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009086 | methionine biosynthesis | 0.00689992 | 1 | Phatr_bicluster_0153 |
| GO:0006094 | gluconeogenesis | 0.00861852 | 1 | Phatr_bicluster_0153 |
| GO:0006099 | tricarboxylic acid cycle | 0.013759 | 1 | Phatr_bicluster_0153 |
| GO:0009396 | folic acid and derivative biosynthesis | 0.013759 | 1 | Phatr_bicluster_0153 |
| GO:0006520 | amino acid metabolism | 0.0407909 | 1 | Phatr_bicluster_0153 |
| GO:0006457 | protein folding | 0.185684 | 1 | Phatr_bicluster_0153 |
| GO:0008152 | metabolism | 0.410263 | 1 | Phatr_bicluster_0153 |

Comments