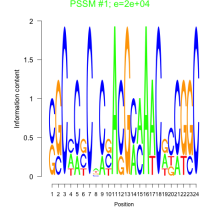
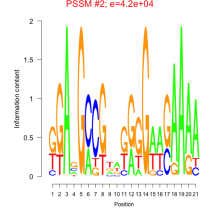
Phatr_bicluster_0154 Residual: 0.20
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0154 | 0.20 | Phaeodactylum tricornutum |
Displaying 1 - 20 of 20
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10310 dna-directed rna polymerase i subunit (Zn-ribbon_RPA12)
PHATRDRAFT_11894 spb1_schpoet-dependent rrna methyltransferase spb1 (2-o-ribose rna methyltransferase) (s-adenosyl-l-methionine-dependent methyltransferase) (AdoMet_MTases)
PHATRDRAFT_15354 rrp5 protein homolog (programmed cell death protein 11)
PHATRDRAFT_15639 mitochondrial carrier protein (Mito_carr)
PHATRDRAFT_15757 small nucleolar ribonucleoprotein u3 component (RpsD)
PHATRDRAFT_16603 mitochondrial translation initiation factor if- (PRK04004)
PHATRDRAFT_16688 cgi-09 protein (Gcd10p superfamily)
PHATRDRAFT_18579 at1g56110 t6h22_9 (SIK1)
PHATRDRAFT_21557 (Brix superfamily)
PHATRDRAFT_23286 3-5 exoribonuclease ph component of the exosome (RNase_PH superfamily)
PHATRDRAFT_25666 periodic tryptophan protein 2 (WD40)
PHATRDRAFT_28984 mgc81303 protein (DEADc)
PHATRDRAFT_30965 radical samcfr familyexpressed (COG0820)
PHATRDRAFT_39052 pentatricopeptiderepeat-containing protein (PPR_2)
PHATRDRAFT_39714 dead (asp-glu-ala-asp) box polypeptide 28 (DEXDc superfamily)
PHATRDRAFT_44096 rna methylase (COG0116)
PHATRDRAFT_44242 u3 snornp protein utp4 (WD40 superfamily)
PHATRDRAFT_47514 elongation protein 4 homolog (PAXNEB superfamily)
PHATRDRAFT_48486 histone tail methylase containing set domain (SET)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006354 | RNA elongation | 0.00739277 | 1 | Phatr_bicluster_0154 |
| GO:0006396 | RNA processing | 0.00787469 | 1 | Phatr_bicluster_0154 |
| GO:0006446 | regulation of translational initiation | 0.00984608 | 1 | Phatr_bicluster_0154 |
| GO:0006306 | DNA methylation | 0.0483639 | 1 | Phatr_bicluster_0154 |
| GO:0006412 | protein biosynthesis | 0.0573335 | 1 | Phatr_bicluster_0154 |
| GO:0006413 | translational initiation | 0.0577801 | 1 | Phatr_bicluster_0154 |
| GO:0006810 | transport | 0.38354 | 1 | Phatr_bicluster_0154 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.426896 | 1 | Phatr_bicluster_0154 |

Comments