Phatr_bicluster_0157 Residual: 0.27
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0157 | 0.27 | Phaeodactylum tricornutum |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11368 pdk1 (3-phosphoinositide-dependent protein kinase 1) kinase (PKc_like superfamily)
PHATRDRAFT_1862 ammonium transporter-like protein amt1 (Ammonium_transp superfamily)
PHATRDRAFT_37052 hsf-type dna-binding domain containing protein (HSF_DNA-bind superfamily)
PHATRDRAFT_37053 kismet cg3696-isoform a
PHATRDRAFT_43631 gene family 7 (TPM)
PHATRDRAFT_45111 yfjf_bacsu upf0060 membrane protein yfjf (PRK02237)
PHATRDRAFT_45163 (Glyco_tranf_2_4)
PHATRDRAFT_46007 (SNF2_N)
PHATRDRAFT_46884 transducin family protein wd-40 repeat family protein
PHATRDRAFT_4918 phosphatidylinositol 3- and 4-kinase family protein ubiquitin family protein (PI3Kc_like superfamily)
PHATRDRAFT_51825 member ras oncogene family (Ras_like_GTPase superfamily)
PHATRDRAFT_54119 (AMN1 superfamily)
PHATRDRAFT_5526 atg1_debha serine threonine-protein kinase atg1 (autophagy-related protein 1) (S_TKc)
PHATRDRAFT_6927 proline betainemfs family protein
PHATRDRAFT_8239 chaperone protein dnaj (DnaJ)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006468 | protein amino acid phosphorylation | 0.0424353 | 1 | Phatr_bicluster_0157 |
| GO:0006810 | transport | 0.0513747 | 1 | Phatr_bicluster_0157 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.0708679 | 1 | Phatr_bicluster_0157 |
| GO:0016567 | protein ubiquitination | 0.13382 | 1 | Phatr_bicluster_0157 |
| GO:0006457 | protein folding | 0.209255 | 1 | Phatr_bicluster_0157 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.359328 | 1 | Phatr_bicluster_0157 |


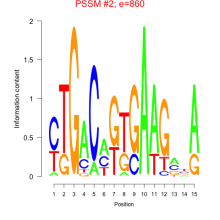
Comments