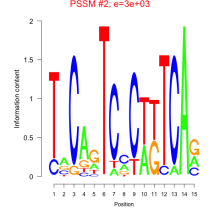
Phatr_bicluster_0184 Residual: 0.27
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0184 | 0.27 | Phaeodactylum tricornutum |
Displaying 1 - 22 of 22
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_13157 small gtp-binding protein domain containing protein (Ras_like_GTPase superfamily)
PHATRDRAFT_36077 tim-barrelyjbn family (DUS_like_FMN)
PHATRDRAFT_37223 (Ten1_2 superfamily)
PHATRDRAFT_38927 gcn5-related n-acetyltransferase
PHATRDRAFT_39329 ribosomal protein l11 methyltransferase
PHATRDRAFT_39520 temporarily assigned gene name family member (tag-30) (BTB)
PHATRDRAFT_42818 xenotropic and polytropic murine leukemia virus receptor xpr1 (EXS superfamily)
PHATRDRAFT_43435 thioredoxin-like proteinexpressed (PITH)
PHATRDRAFT_43454 cog0457: fog: tpr repeat (COG4976)
PHATRDRAFT_44453 solute carrier family 29 (nucleoside transporters)memberisoform cra_b (Nucleoside_tran superfamily)
PHATRDRAFT_45790 myeloid lymphoid or mixed-lineage leukemia (zf-CXXC)
PHATRDRAFT_46920 (acyl_WS_DGAT)
PHATRDRAFT_47117 p-atpase family transporter: cadmium zinc ion heavy metal translocating p-type atpase family-like protein
PHATRDRAFT_47603 (Gly_transf_sug superfamily)
PHATRDRAFT_48569 abortive infection protein (Abi)
PHATRDRAFT_54834 (CpeT superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006886 | intracellular protein transport | 0.0389484 | 1 | Phatr_bicluster_0184 |
| GO:0015031 | protein transport | 0.030286 | 1 | Phatr_bicluster_0184 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.199489 | 1 | Phatr_bicluster_0184 |
| GO:0007264 | small GTPase mediated signal transduction | 0.0205926 | 1 | Phatr_bicluster_0184 |

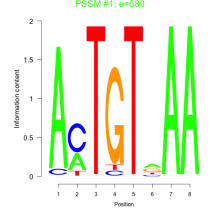
Comments