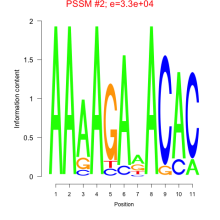
Phatr_bicluster_0193 Residual: 0.36
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0193 | 0.36 | Phaeodactylum tricornutum |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11901 chromatin modifying protein 1b (Snf7 superfamily)
PHATRDRAFT_15749 leucine rich repeat-like protein
PHATRDRAFT_42443 wd-40 repeat (WD40 superfamily)
PHATRDRAFT_44873 protein epd2 precursor (Glyco_hydro_72 superfamily)
PHATRDRAFT_46213 ganp protein (SAC3_GANP)
PHATRDRAFT_47686 (Alginate_lyase)
PHATRDRAFT_50133 smp-30 gluconolaconase lre-like region (SGL)
PHATRDRAFT_50250 (ANK)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006813 | potassium ion transport | 0.014749 | 1 | Phatr_bicluster_0193 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.15368 | 1 | Phatr_bicluster_0193 |
| GO:0008152 | metabolism | 0.202444 | 1 | Phatr_bicluster_0193 |

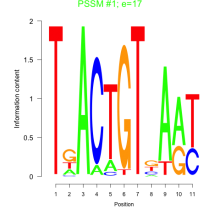
Comments