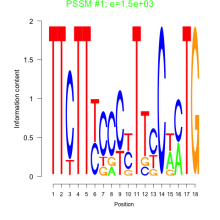
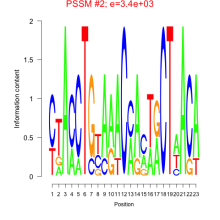
Phatr_bicluster_0200 Residual: 0.32
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0200 | 0.32 | Phaeodactylum tricornutum |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10835 translation initiation factor if-2 (IF-2)
PHATRDRAFT_12679 mgc140358 protein (Brix)
PHATRDRAFT_13235 dead (asp-glu-ala-asp) box polypeptide 10 (DEADc)
PHATRDRAFT_16592 peroxisomal membrane protein 22 (Mpv17_PMP22)
PHATRDRAFT_16633 rna-binding protein (RRM_1)
PHATRDRAFT_19579 glucose inhibited division protein a (PRK05192)
PHATRDRAFT_35057 indole-3-glycerol-phosphate synthase (TIM_phosphate_binding superfamily)
PHATRDRAFT_37260 mitochondrial rna splicing protein (Mrs2_Mfm1p-like)
PHATRDRAFT_37681 eukaryotic translation initiation factor 3 subunit (SAP)
PHATRDRAFT_38628 membrane protein
PHATRDRAFT_43482 serine threonine protein kinase (MPP_PPP_family)
PHATRDRAFT_43627 (IKI3)
PHATRDRAFT_44863 polyribonucleotide nucleotidyltransferase (PRK11824)
PHATRDRAFT_45529 ribonucleoside-diphosphate reductase large subunit (PLN02437)
PHATRDRAFT_46041 dna binding nuclease (SnoaL_3)
PHATRDRAFT_46130 mgc80131 protein (zf-MYND)
PHATRDRAFT_46782 alkylated dna repair protein alkb homolog (2OG-FeII_Oxy_2)
PHATRDRAFT_47890 y3488_glovi upf0090 protein glr3488 (Sm_like superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009231 | riboflavin biosynthesis | 0.0103346 | 1 | Phatr_bicluster_0200 |
| GO:0006568 | tryptophan metabolism | 0.0103346 | 1 | Phatr_bicluster_0200 |
| GO:0030001 | metal ion transport | 0.0273554 | 1 | Phatr_bicluster_0200 |
| GO:0006413 | translational initiation | 0.0407909 | 1 | Phatr_bicluster_0200 |
| GO:0006260 | DNA replication | 0.0441247 | 1 | Phatr_bicluster_0200 |
| GO:0006396 | RNA processing | 0.0929544 | 1 | Phatr_bicluster_0200 |
| GO:0006412 | protein biosynthesis | 0.247414 | 1 | Phatr_bicluster_0200 |

Comments