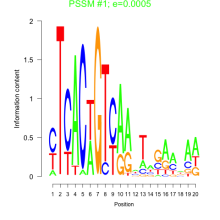
Phatr_bicluster_0202 Residual: 0.37
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0202 | 0.37 | Phaeodactylum tricornutum |
Displaying 1 - 32 of 32
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_14067 trna-pseudouridine synthase (PseudoU_synth superfamily)
PHATRDRAFT_16261 disease resistance protein
PHATRDRAFT_33313 ankyrin-repeat protein ank-1 (ANK)
PHATRDRAFT_38427 2og-feoxygenase family (2OG-FeII_Oxy_3)
PHATRDRAFT_43903 conserved hypothetical membrane protein
PHATRDRAFT_44111 mopfamily transporter (MATE_like superfamily)
PHATRDRAFT_46264 mgc68579 protein (HATPase_c)
PHATRDRAFT_49136 heat shock transcription factor hsf1 (HSF_DNA-bind)
PHATRDRAFT_49312 (SPS1)
PHATRDRAFT_50253 apg4-d protein (Peptidase_C54 superfamily)
PHATRDRAFT_50468 (Sulfotransfer_2 superfamily)
PHATRDRAFT_50508 conserved hypothetical membrane protein (DUF4395)
PHATRDRAFT_55091 cryptochrome dash (crypto_DASH)
PHATRDRAFT_6914 peptide deformylase (Pep_deformylase)
PHATRDRAFT_9478 phytochelatin synthase (Phytochelatin)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006396 | RNA processing | 0.0802144 | 1 | Phatr_bicluster_0202 |
| GO:0006412 | protein biosynthesis | 0.216202 | 1 | Phatr_bicluster_0202 |
| GO:0006468 | protein amino acid phosphorylation | 0.229422 | 1 | Phatr_bicluster_0202 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.283834 | 1 | Phatr_bicluster_0202 |
| GO:0006118 | electron transport | 0.294987 | 1 | Phatr_bicluster_0202 |
| GO:0019538 | protein metabolism | 0.0522062 | 1 | Phatr_bicluster_0202 |


Comments