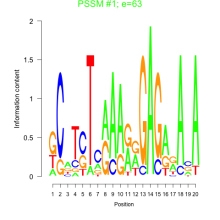
Phatr_bicluster_0203 Residual: 0.15
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0203 | 0.15 | Phaeodactylum tricornutum |
Displaying 1 - 21 of 21
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_38462 copia protein (gag-int-pol protein)
PHATRDRAFT_39129 multicoppertype 2 (Cupredoxin superfamily)
PHATRDRAFT_39223 retroelement pol polyprotein
PHATRDRAFT_39341 gag-pol polyprotein (rve)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006310 | DNA recombination | 0.000798705 | 1 | Phatr_bicluster_0203 |
| GO:0006508 | proteolysis and peptidolysis | 0.0272679 | 1 | Phatr_bicluster_0203 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.0448846 | 1 | Phatr_bicluster_0203 |


Comments