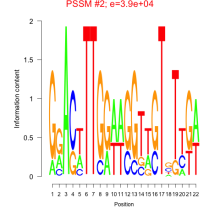
Phatr_bicluster_0208 Residual: 0.30
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0208 | 0.30 | Phaeodactylum tricornutum |
Displaying 1 - 22 of 22
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11236 dna repair protein (PRK11823)
PHATRDRAFT_15042 topoisomerase 6 subunit b (top6b)
PHATRDRAFT_3171 mitochondrial translocase complex component (NIF)
PHATRDRAFT_36197 soul3-like protein
PHATRDRAFT_37127 membrane bound o-acyl transferasefamily protein wax synthase-related (MBOAT_2)
PHATRDRAFT_42744 (FSH1 superfamily)
PHATRDRAFT_42864 httm domain protein (HTTM superfamily)
PHATRDRAFT_43425 salicylate hydroxylase (UbiH)
PHATRDRAFT_43598 kelch motif family protein
PHATRDRAFT_44801 possible acetyltransferasefamily protein
PHATRDRAFT_45766 (DUF3054)
PHATRDRAFT_46706 biotin lipoate protein ligase-like protein (PTZ00276)
PHATRDRAFT_48762 oxidoreductase family protein (MviM)
PHATRDRAFT_49372 tha1 (threonine aldolase 1) aldehyde-lyase (PLN02721)
PHATRDRAFT_8940 fkbp-type peptidyl-prolyl cis-trans isomerase (FKBP_C)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006725 | aromatic compound metabolism | 0.0388725 | 1 | Phatr_bicluster_0208 |
| GO:0006118 | electron transport | 0.3726 | 1 | Phatr_bicluster_0208 |
| GO:0008152 | metabolism | 0.453158 | 1 | Phatr_bicluster_0208 |
| GO:0006813 | potassium ion transport | 0.0388725 | 1 | Phatr_bicluster_0208 |
| GO:0006457 | protein folding | 0.209255 | 1 | Phatr_bicluster_0208 |
| GO:0006464 | protein modification | 0.118 | 1 | Phatr_bicluster_0208 |
| GO:0006508 | proteolysis and peptidolysis | 0.365994 | 1 | Phatr_bicluster_0208 |
| GO:0006810 | transport | 0.320848 | 1 | Phatr_bicluster_0208 |

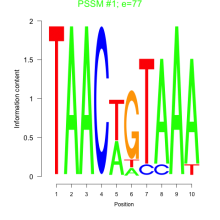
Comments