Phatr_bicluster_0210 Residual: 0.20
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0210 | 0.20 | Phaeodactylum tricornutum |
Displaying 1 - 20 of 20
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10757 aspartate-semialdehyde dehydrogenase (PLN02383)
PHATRDRAFT_12254 fkbp-type peptidyl-prolyl cis-trans isomerase (FkpA)
PHATRDRAFT_12963 (ATP_bind_1)
PHATRDRAFT_13119 atp-binding family protein (ATP_bind_1)
PHATRDRAFT_13174 ascorbate peroxidase (ascorbate_peroxidase)
PHATRDRAFT_19413 drosophila melanogaster cg18001 (Ribosomal_L38e)
PHATRDRAFT_21929 (Coatomer_WDAD)
PHATRDRAFT_27851 40s ribosomal protein s10 (S10_plectin)
PHATRDRAFT_28147 40s ribosomal protein s29-a (Ribosomal_S14 superfamily)
PHATRDRAFT_33660 40s ribosomal protein (PTZ00084)
PHATRDRAFT_34526 argininosuccinate lyase (PLN02646)
PHATRDRAFT_36226 ribosomal protein l18a (RPL20A)
PHATRDRAFT_36420 lsu ribosomal protein l9p (Ribosomal_L9_N)
PHATRDRAFT_41252 (OTU)
PHATRDRAFT_42104 nucleoside diphosphate kinase (NDPk_I)
PHATRDRAFT_44291 ribosomal protein (Ribosomal_L34e superfamily)
PHATRDRAFT_50701 yeast ribosomal protein s28 homologue (Ribosomal_S23)
PHATRDRAFT_8058 ankyrin repeat protein mbp3_16 (ANK)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006412 | protein biosynthesis | 0.0000207 | 0.0479151 | Phatr_bicluster_0210 |
| GO:0006241 | CTP biosynthesis | 0.0118109 | 1 | Phatr_bicluster_0210 |
| GO:0006183 | GTP biosynthesis | 0.0118109 | 1 | Phatr_bicluster_0210 |
| GO:0006228 | UTP biosynthesis | 0.0118109 | 1 | Phatr_bicluster_0210 |
| GO:0009088 | threonine biosynthesis | 0.0118109 | 1 | Phatr_bicluster_0210 |
| GO:0009086 | methionine biosynthesis | 0.0157187 | 1 | Phatr_bicluster_0210 |
| GO:0006414 | translational elongation | 0.0540081 | 1 | Phatr_bicluster_0210 |
| GO:0006979 | response to oxidative stress | 0.0908976 | 1 | Phatr_bicluster_0210 |
| GO:0006520 | amino acid metabolism | 0.0908976 | 1 | Phatr_bicluster_0210 |
| GO:0006886 | intracellular protein transport | 0.147128 | 1 | Phatr_bicluster_0210 |

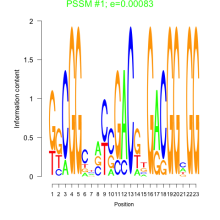
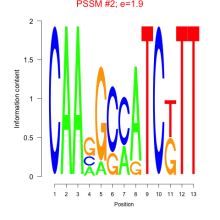
Comments