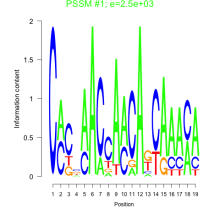
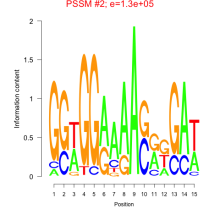
Phatr_bicluster_0215 Residual: 0.22
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0215 | 0.22 | Phaeodactylum tricornutum |
Displaying 1 - 29 of 29
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10782 rna helicase (DEADc)
PHATRDRAFT_12779 dna-directed rna polymerases i and iii 16 kda polypeptide (RNAP_I_III_AC19)
PHATRDRAFT_13428 (WD40 superfamily)
PHATRDRAFT_13820 bystin family protein (Bystin)
PHATRDRAFT_17523 ribonucleoside-diphosphatebeta subunit family protein (RNRR2)
PHATRDRAFT_17974 rnu3ip2 protein (WD40 superfamily)
PHATRDRAFT_18217 mitogen-activated protein kinase organizer 1 (WD40)
PHATRDRAFT_25394 loc100036868 protein (nop2p)
PHATRDRAFT_32036 noc2l_arath nucleolar complex protein 2 homolog (protein noc2 homolog) (Noc2 superfamily)
PHATRDRAFT_35048 polybindingnuclear 1 (RRM_SF superfamily)
PHATRDRAFT_35267 (RBFA superfamily)
PHATRDRAFT_35966 nop seven associated protein
PHATRDRAFT_40101 corneal wound healing-related protein (Mak10 superfamily)
PHATRDRAFT_43703 af332588_1cell growth-regulating nucleolar protein
PHATRDRAFT_45441 mis3_schpo ribosomal rna assembly protein mis3 (COG1094)
PHATRDRAFT_45727 mak5_emeni atp-dependent rna helicase mak5 (SrmB)
PHATRDRAFT_45844 at1g04130 f20d22_10 (TPR)
PHATRDRAFT_46259 rna-dependent helicase (DEADc)
PHATRDRAFT_46848 (COG2520)
PHATRDRAFT_47198 receptor for activated c (WD40 superfamily)
PHATRDRAFT_51897 nucleolar protein 10 (WD40 superfamily)
PHATRDRAFT_6258 af155362_1charged amino acid rich leucine zipper factor-1 (Sas10)
PHATRDRAFT_8777 af361826_1 at1g42440 f7f22_7 (DUF663)
PHATRDRAFT_9929 at3g22660 mwi23_3 (Ebp2 superfamily)
PHATRDRAFT_9989 putatative 28 kda protein (COG1094)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009186 | deoxyribonucleoside diphosphate metabolism | 0.00370096 | 1 | Phatr_bicluster_0215 |
| GO:0006364 | rRNA processing | 0.00984487 | 1 | Phatr_bicluster_0215 |
| GO:0006350 | transcription | 0.0256773 | 1 | Phatr_bicluster_0215 |
| GO:0005975 | carbohydrate metabolism | 0.0754495 | 1 | Phatr_bicluster_0215 |
| GO:0006810 | transport | 0.21473 | 1 | Phatr_bicluster_0215 |

Comments