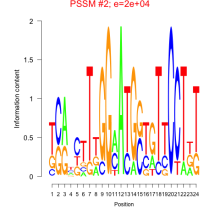
Phatr_bicluster_0220 Residual: 0.29
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0220 | 0.29 | Phaeodactylum tricornutum |
Displaying 1 - 29 of 29
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10068 enoyl-acyl carrier reductase (PLN02730)
PHATRDRAFT_10896 solute carrier protein (Mito_carr)
PHATRDRAFT_13895 photosystem ii stability assembly factor hcf136 (PLN00033)
PHATRDRAFT_14529 inorganic pyrophosphatase-like protein (pyrophosphatase)
PHATRDRAFT_20657 chloroplast atpase gamma subunit precursor (F1-ATPase_gamma)
PHATRDRAFT_22122 glyceraldehyde-3-phosphate dehydrogenase precursor (GapA)
PHATRDRAFT_32708 (PLN02307)
PHATRDRAFT_35386 methyltransferase type 11 (Methyltransf_12)
PHATRDRAFT_35587 mopfamily transporter: multidrug efflux (MATE_like superfamily)
PHATRDRAFT_41038 glutaredoxin 2 (PRK10387)
PHATRDRAFT_43233 atp sulfurylase (sulfate adenylyltransferase) (sopT)
PHATRDRAFT_44603 low-co2 inducible protein
PHATRDRAFT_44999 atp synthase h+-transporting mitochondrial f1 complex o subunit (OSCP)
PHATRDRAFT_45190 (alpha-crystallin-Hsps_p23-like superfamily)
PHATRDRAFT_45465 gnat family (RimI)
PHATRDRAFT_46930 deoxyxylulose-5-phosphate synthase (TIP120 superfamily)
PHATRDRAFT_47103 (Methyltransf_16 superfamily)
PHATRDRAFT_49779 10-formyltetrahydrofolate synthetase (FTHFS)
PHATRDRAFT_8747 glutaredoxin type i (Thioredoxin_like superfamily)
PHATRDRAFT_9709 (COG1739)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015986 | ATP synthesis coupled proton transport | 0.00690102 | 1 | Phatr_bicluster_0220 |
| GO:0000103 | sulfate assimilation | 0.00690418 | 1 | Phatr_bicluster_0220 |
| GO:0006855 | multidrug transport | 0.0137641 | 1 | Phatr_bicluster_0220 |
| GO:0045454 | cell redox homeostasis | 0.02058 | 1 | Phatr_bicluster_0220 |
| GO:0006006 | glucose metabolism | 0.0239715 | 1 | Phatr_bicluster_0220 |
| GO:0009396 | folic acid and derivative biosynthesis | 0.0273521 | 1 | Phatr_bicluster_0220 |
| GO:0006633 | fatty acid biosynthesis | 0.0440926 | 1 | Phatr_bicluster_0220 |
| GO:0006096 | glycolysis | 0.114621 | 1 | Phatr_bicluster_0220 |
| GO:0005975 | carbohydrate metabolism | 0.197401 | 1 | Phatr_bicluster_0220 |
| GO:0008152 | metabolism | 0.270353 | 1 | Phatr_bicluster_0220 |

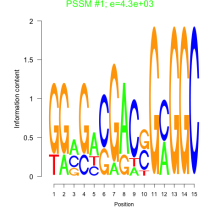
Comments