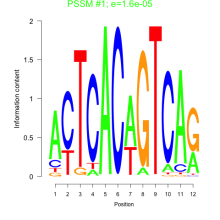
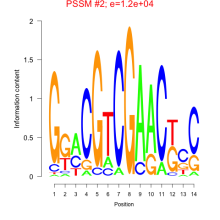
Phatr_bicluster_0222 Residual: 0.32
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0222 | 0.32 | Phaeodactylum tricornutum |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11673 band 7 family protein (SPFH_like_u4)
PHATRDRAFT_28743 wdr82 protein (WD40 superfamily)
PHATRDRAFT_33651 cral trio cell signalling protein (SEC14)
PHATRDRAFT_36891 (R3H superfamily)
PHATRDRAFT_41659 atp-dependent transporter (Uup)
PHATRDRAFT_42948 viral a-type inclusion
PHATRDRAFT_43226 n-terminal domain containing protein
PHATRDRAFT_44351 mpt family transporter: inner membrane translocasetim44 (Tim44 superfamily)
PHATRDRAFT_44427 glucose-repressible alcohol dehydrogenase transcriptional effector ccr4 and related proteins (EEP superfamily)
PHATRDRAFT_44473 deacetylase sulfotransferase
PHATRDRAFT_44526 (alpha_CA superfamily)
PHATRDRAFT_48720 ccr4 associated factor-like protein
PHATRDRAFT_48786 tpr-repeat containing protein (TPR)
PHATRDRAFT_49026 (Glyco_trans_2_3)
PHATRDRAFT_49385 deoxycytidyl transferase (PolY superfamily)
PHATRDRAFT_54411 sugar (d-ribose) abc transporter (periplasmic binding protein) (Peripla_BP_4)
PHATRDRAFT_708 yk16_schpoatp-dependent rna helicase (HrpA)
PHATRDRAFT_7114 tim22_yarli mitochondrial import inner membrane translocase subunit tim22 (Tim17)
PHATRDRAFT_8982 phosphoglycerate mutase (HP_PGM_like)
PHATRDRAFT_9879 haloacid dehalogenase-like hydrolase (HAD superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006730 | one-carbon compound metabolism | 0.00886476 | 1 | Phatr_bicluster_0222 |
| GO:0015031 | protein transport | 0.066897 | 1 | Phatr_bicluster_0222 |
| GO:0006281 | DNA repair | 0.0731548 | 1 | Phatr_bicluster_0222 |
| GO:0000074 | regulation of cell cycle | 0.0731548 | 1 | Phatr_bicluster_0222 |
| GO:0006096 | glycolysis | 0.0752324 | 1 | Phatr_bicluster_0222 |
| GO:0006886 | intracellular protein transport | 0.0855585 | 1 | Phatr_bicluster_0222 |
| GO:0006396 | RNA processing | 0.117917 | 1 | Phatr_bicluster_0222 |
| GO:0006810 | transport | 0.352949 | 1 | Phatr_bicluster_0222 |
| GO:0008152 | metabolism | 0.492943 | 1 | Phatr_bicluster_0222 |

Comments