Phatr_bicluster_0223 Residual: 0.26
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0223 | 0.26 | Phaeodactylum tricornutum |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12668 ribulose-phosphate 3-epimerase (RPE)
PHATRDRAFT_12881 carbohydrate kinase fggy (NBD_sugar-kinase_HSP70_actin superfamily)
PHATRDRAFT_20192 d-lactate dehydrogenase (GlcD)
PHATRDRAFT_2793 chloroplast fructose--bisphosphatase (FBPase)
PHATRDRAFT_37212 sac3 ganp domain protein (SAC3_GANP)
PHATRDRAFT_37749 protein n-methyltransferase
PHATRDRAFT_42595 phd finger protein 8 (JmjC superfamily)
PHATRDRAFT_42694 (DUF4205 superfamily)
PHATRDRAFT_42834 kelch motif family protein (BTB)
PHATRDRAFT_43063 (DUF3082 superfamily)
PHATRDRAFT_43165 protein tyrosinemitochondrial 1 (CDC14)
PHATRDRAFT_43727 (DUF599 superfamily)
PHATRDRAFT_43747 zincmynd-type containing 15 isoform 1
PHATRDRAFT_44398 abc transporter g family protein (ABC_ATPase)
PHATRDRAFT_45364 wd-repeat protein (WD40 superfamily)
PHATRDRAFT_46737 vad1 (vascular associated death1)
PHATRDRAFT_49206 (NCA2 superfamily)
PHATRDRAFT_50961 mitochondrial pyruvate dehydrogenase kinase (BCDHK_Adom3)
PHATRDRAFT_6635 member ras oncogene family like (Ras_like_GTPase superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0005975 | carbohydrate metabolism | 0.000282174 | 0.64646 | Phatr_bicluster_0223 |
| GO:0016310 | phosphorylation | 0.0307218 | 1 | Phatr_bicluster_0223 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0414888 | 1 | Phatr_bicluster_0223 |
| GO:0007264 | small GTPase mediated signal transduction | 0.0457657 | 1 | Phatr_bicluster_0223 |
| GO:0015031 | protein transport | 0.066897 | 1 | Phatr_bicluster_0223 |
| GO:0006468 | protein amino acid phosphorylation | 0.323667 | 1 | Phatr_bicluster_0223 |
| GO:0006118 | electron transport | 0.408154 | 1 | Phatr_bicluster_0223 |

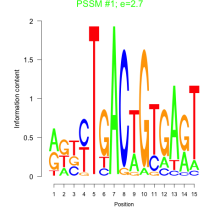

Comments