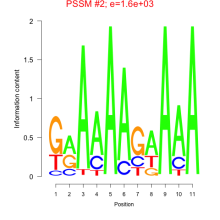
Phatr_bicluster_0229 Residual: 0.31
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0229 | 0.31 | Phaeodactylum tricornutum |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_13331 peptidase s41a (prc)
PHATRDRAFT_14064 protein kinase c inhibitor (PKCI_related)
PHATRDRAFT_17401 dihydrolipoamide s-acetyltransferase (e2 component of pyruvate dehydrogenase complex)isoform cra_a (PDHac_trf_mito)
PHATRDRAFT_20183 at5g50850 k16e14_1 (PLN02683)
PHATRDRAFT_21122 beta-tubulin (PLN00220)
PHATRDRAFT_28730 peptidyl-prolyl cis-trans isomerase (PTZ00356)
PHATRDRAFT_35063 snrnp protein (Sm_B)
PHATRDRAFT_35580 prune homolog (PPX1)
PHATRDRAFT_39252 ubiquitin family protein (UBL)
PHATRDRAFT_42545 mg2+ transporter (MgtE)
PHATRDRAFT_44204 cog2940: proteins containing set domain (SET)
PHATRDRAFT_44425 asparagine-linked glycosylation 12 homolog (alpha--mannosyltransferase) (Glyco_transf_22 superfamily)
PHATRDRAFT_44639 abc(binding protein) family transporter: phosphate (Phosphonate-bd superfamily)
PHATRDRAFT_46613 xanthine phosphoribosyltransferase (PRTases_typeI)
PHATRDRAFT_46710 proteophosphoglycan 5
PHATRDRAFT_49630 (EMP24_GP25L)
PHATRDRAFT_50148 protein phosphatase 2c (PP2Cc)
PHATRDRAFT_54086 atp synthase beta subunit (PRK09280)
PHATRDRAFT_54730 beta-adaptin-like protein a (Adaptin_N)
PHATRDRAFT_9348 dcmp deaminase isoform 3 (deoxycytidylate_deaminase)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0043101 | purine salvage | 0.00271672 | 1 | Phatr_bicluster_0229 |
| GO:0046110 | xanthine metabolism | 0.00271672 | 1 | Phatr_bicluster_0229 |
| GO:0051258 | protein polymerization | 0.0135166 | 1 | Phatr_bicluster_0229 |
| GO:0009116 | nucleoside metabolism | 0.0242101 | 1 | Phatr_bicluster_0229 |
| GO:0006397 | mRNA processing | 0.0608128 | 1 | Phatr_bicluster_0229 |
| GO:0006812 | cation transport | 0.0710389 | 1 | Phatr_bicluster_0229 |
| GO:0007018 | microtubule-based movement | 0.078642 | 1 | Phatr_bicluster_0229 |
| GO:0006464 | protein modification | 0.158622 | 1 | Phatr_bicluster_0229 |
| GO:0006810 | transport | 0.412691 | 1 | Phatr_bicluster_0229 |
| GO:0006508 | proteolysis and peptidolysis | 0.465714 | 1 | Phatr_bicluster_0229 |


Comments