Phatr_bicluster_0230 Residual: 0.25
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0230 | 0.25 | Phaeodactylum tricornutum |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11151 dihydrodipicolinate synthase (DHDPS)
PHATRDRAFT_11740 orotate phosphoribosyltransferase (PRTases_typeI)
PHATRDRAFT_15019 rrna methylase family protein (SpoU)
PHATRDRAFT_16281 mgc83338 protein (RRM_eIF3G_like)
PHATRDRAFT_17779 nucleosome assembly protein (NAP)
PHATRDRAFT_20589 diptheria toxin resistance protein required for diphthamide biosynthesis-like 1 (diphth2_R)
PHATRDRAFT_24408 mgc154353 protein (RimI)
PHATRDRAFT_25204 phosphoribosylaminoimidazole carboxylase (PLN02948)
PHATRDRAFT_31975 protein phosphatasecatalytic subunit (MPP_PP5_C)
PHATRDRAFT_33523 wd-40 repeat family protein (WD40 superfamily)
PHATRDRAFT_3578 plastid ribosomal protein l21 large ribosomal subunit (rplU)
PHATRDRAFT_36743 pseudouridylate synthase (PseudoU_synth_PUS1_PUS2)
PHATRDRAFT_38881 (Ribosomal_L7Ae)
PHATRDRAFT_42495 tryptophanyl-trna synthetase (TrpRS_core)
PHATRDRAFT_43016 (Utp21 superfamily)
PHATRDRAFT_43135 valyl trna synthetase (PTZ00419)
PHATRDRAFT_44446 rh26_orysj dead-box atp-dependent rna helicase 26 (DEXDc superfamily)
PHATRDRAFT_45520 fertility restorer
PHATRDRAFT_45976 (PPR_2)
PHATRDRAFT_48430 transportin 2 (importinkaryopherin beta 2b) (KAP95)
PHATRDRAFT_51303 signal recognition particle 68 kda protein
PHATRDRAFT_52094 (RRM_SF superfamily)
PHATRDRAFT_8842 rna polymerase i subunit hrpa39 (RNAP_I_II_AC40)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006189 | 'de novo' IMP biosynthesis | 0.0159584 | 1 | Phatr_bicluster_0230 |
| GO:0009116 | nucleoside metabolism | 0.0285555 | 1 | Phatr_bicluster_0230 |
| GO:0006334 | nucleosome assembly | 0.0502429 | 1 | Phatr_bicluster_0230 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0594 | 1 | Phatr_bicluster_0230 |
| GO:0006412 | protein biosynthesis | 0.410379 | 1 | Phatr_bicluster_0230 |
| GO:0006221 | pyrimidine nucleotide biosynthesis | 0.00641182 | 1 | Phatr_bicluster_0230 |
| GO:0042254 | ribosome biogenesis and assembly | 0.0191218 | 1 | Phatr_bicluster_0230 |
| GO:0006396 | RNA processing | 0.165834 | 1 | Phatr_bicluster_0230 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.0115276 | 1 | Phatr_bicluster_0230 |
| GO:0008033 | tRNA processing | 0.0624342 | 1 | Phatr_bicluster_0230 |


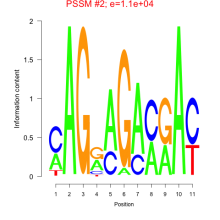
Comments